6YC3
 
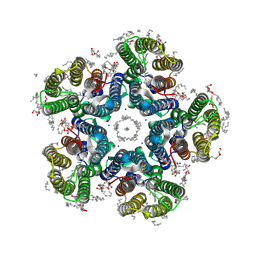 | | Crystal structure of the light-driven sodium pump KR2 in the pentameric form, pH 8.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Gushchin, I, Gordeliy, V. | | Deposit date: | 2020-03-18 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular mechanism of light-driven sodium pumping.
Nat Commun, 11, 2020
|
|
6SUM
 
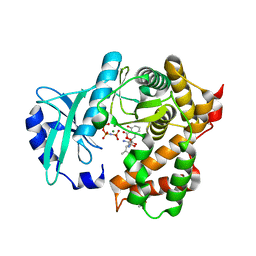 | | Amicoumacin kinase hAmiN in complex with AMP-PNP, MG2+ and Ami | | Descriptor: | ACETATE ION, AMICOUMACIN KINASE, Amicoumacin A, ... | | Authors: | Bourenkov, G.P, Mokrushina, Y.A, Terekhov, S.S, Smirnov, I.V, Gabibov, A.G, Altman, S. | | Deposit date: | 2019-09-16 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A kinase bioscavenger provides antibiotic resistance by extremely tight substrate binding.
Sci Adv, 6, 2020
|
|
6SUL
 
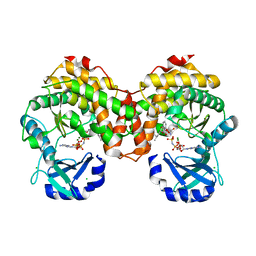 | | Amicoumacin kinase AmiN in complex with AMP-PNP, Mg2+ and Ami | | Descriptor: | Amicoumacin A, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bourenkov, G.P, Mokrushina, Y.A, Terekhov, S.S, Smirnov, I.V, Gabibov, A.G, Altman, S. | | Deposit date: | 2019-09-15 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A kinase bioscavenger provides antibiotic resistance by extremely tight substrate binding.
Sci Adv, 6, 2020
|
|
6YC4
 
 | | Crystal structure of the steady-state activated state of the light-driven sodium pump KR2 in the pentameric form at room temperature, pH 8.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, GLYCEROL, ... | | Authors: | Kovalev, K, Gushchin, I, Gordeliy, V. | | Deposit date: | 2020-03-18 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular mechanism of light-driven sodium pumping.
Nat Commun, 11, 2020
|
|
6YBY
 
 | | Crystal structure of the D116N mutant of the light-driven sodium pump KR2 in the monomeric form, pH 4.6 | | Descriptor: | EICOSANE, RETINAL, SODIUM ION, ... | | Authors: | Kovalev, K, Gushchin, I, Gordeliy, V. | | Deposit date: | 2020-03-18 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanism of light-driven sodium pumping.
Nat Commun, 11, 2020
|
|
6YC1
 
 | | Crystal structure of the H30A mutant of the light-driven sodium pump KR2 in the pentameric form, pH 8.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Gushchin, I, Gordeliy, V. | | Deposit date: | 2020-03-18 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular mechanism of light-driven sodium pumping.
Nat Commun, 11, 2020
|
|
6YC0
 
 | | Crystal structure of the steady-state-SMX activated state of the light-driven sodium pump KR2 in the pentameric form at room temperature, pH 8.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, GLYCEROL, ... | | Authors: | Kovalev, K, Gushchin, I, Gordeliy, V. | | Deposit date: | 2020-03-18 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular mechanism of light-driven sodium pumping.
Nat Commun, 11, 2020
|
|
6YC2
 
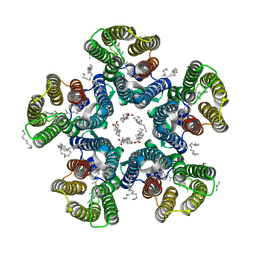 | | Crystal structure of the light-driven sodium pump KR2 in the pentameric form at room temperature, pH 8.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, ALANINE, EICOSANE, ... | | Authors: | Kovalev, K, Gushchin, I, Gordeliy, V. | | Deposit date: | 2020-03-18 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism of light-driven sodium pumping.
Nat Commun, 11, 2020
|
|
6SUI
 
 | | AMICOUMACIN KINASE AMIN | | Descriptor: | PENTAETHYLENE GLYCOL, Phosphotransferase enzyme family protein | | Authors: | Bourenkov, G.P, Mokrushina, Y.A, Terekhov, S.S, Smirnov, I.V, Gabibov, A.G, Altman, S. | | Deposit date: | 2019-09-14 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A kinase bioscavenger provides antibiotic resistance by extremely tight substrate binding.
Sci Adv, 6, 2020
|
|
6SUN
 
 | | Amicoumacin kinase hAmiN in complex with AMP-PNP, Ca2+ and Ami | | Descriptor: | APH domain-containing protein, amicoumacin kinase, Amicoumacin A, ... | | Authors: | Bourenkov, G.P, Mokrushina, Y.A, Terekhov, S.S, Smirnov, I.V, Gabibov, A.G, Altman, S. | | Deposit date: | 2019-09-16 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A kinase bioscavenger provides antibiotic resistance by extremely tight substrate binding.
Sci Adv, 6, 2020
|
|
6SV5
 
 | | Amicoumacin kinase AmiN in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Phosphotransferase enzyme family protein, amicoumacin kinase | | Authors: | Bourenkov, G.P, Mokrushina, Y.A, Terekhov, S.S, Smirnov, I.V, Gabibov, A.G, Altman, S. | | Deposit date: | 2019-09-17 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A kinase bioscavenger provides antibiotic resistance by extremely tight substrate binding.
Sci Adv, 6, 2020
|
|
7OQM
 
 | | Human OMPD-domain of UMPS in complex with substrate OMP at 1.05 Angstroms resolution, 20 minutes soaking | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Rindfleisch, S, Rabe von Pappenheim, F, Tittmann, K. | | Deposit date: | 2021-06-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
7OQN
 
 | | Human OMPD-domain of UMPS in complex with substrate OMP at 1.10 Angstroms resolution, 30 minutes soaking | | Descriptor: | GLYCEROL, SULFATE ION, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Rindfleisch, S, Rabe von Pappenheim, F, Tittmann, K. | | Deposit date: | 2021-06-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
7OUZ
 
 | |
7OV0
 
 | |
7OQF
 
 | | Human OMPD-domain of UMPS in complex with OMP at 1.05 Angstrom resolution, 5 minutes soaking | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Rindfleisch, S, Rabe von Pappenheim, F, Tittmann, K. | | Deposit date: | 2021-06-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
7OQI
 
 | | Human OMPD-domain of UMPS in complex with substrate OMP at 1.15 Angstrom resolution, 10 minutes soaking | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Rindfleisch, S, Rabe von Pappenheim, F, Tittmann, K. | | Deposit date: | 2021-06-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
7OQK
 
 | | Human OMPD-domain of UMPS in complex with substrate OMP at 1.10 Angstroms resolution, 15 minutes soaking | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Rindfleisch, S, Rabe von Pappenheim, F, Tittmann, K. | | Deposit date: | 2021-06-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
7OTU
 
 | | Human OMPD-domain of UMPS in complex with 6-hydroxy-UMP at 0.95 Angstroms resolution, crystal 2 | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Isoform 2 of Uridine 5'-monophosphate synthase | | Authors: | Rindfleisch, S, Rabe von Pappenheim, F, Tittmann, K. | | Deposit date: | 2021-06-10 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
2NAP
 
 | | DISSIMILATORY NITRATE REDUCTASE (NAP) FROM DESULFOVIBRIO DESULFURICANS | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Dias, J.M, Than, M, Humm, A, Huber, R, Bourenkov, G, Bartunik, H, Bursakov, S, Calvete, J, Caldeira, J, Carneiro, C, Moura, J, Moura, I, Romao, M.J. | | Deposit date: | 1998-09-18 | | Release date: | 1999-09-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the first dissimilatory nitrate reductase at 1.9 A solved by MAD methods.
Structure Fold.Des., 7, 1999
|
|
7Q1H
 
 | |
7AQE
 
 | | Structure of SARS-CoV-2 Main Protease bound to UNC-2327 | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P, Meents, A. | | Deposit date: | 2020-10-21 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
4U4M
 
 | | Crystal structure of 0.5M urea unfolded YagE, a KDG aldolase protein in complex with Pyruvate | | Descriptor: | 1,2-ETHANEDIOL, PYRUVIC ACID, UREA, ... | | Authors: | Manoj Kumar, P, Bhaskar, V, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2014-07-24 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Crystal structure of 0.5M urea unfolded YagE, a KDG aldolase protein in complex with Pyruvate
To be published
|
|
1VFB
 
 | |
1VFA
 
 | |
