7CS7
 
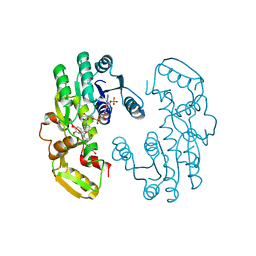 | | IiPLR1 with NADP+ and (+)secoisolariciresinol | | Descriptor: | (2S,3S)-2,3-bis[(3-methoxy-4-oxidanyl-phenyl)methyl]butane-1,4-diol, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol-lariciresinol reductase | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.297653 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CSG
 
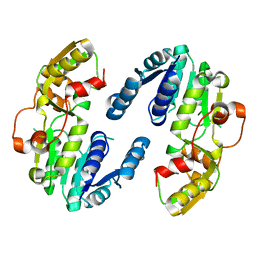 | | AtPrR2 in apo form | | Descriptor: | Pinoresinol reductase 2 | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99689388 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CSA
 
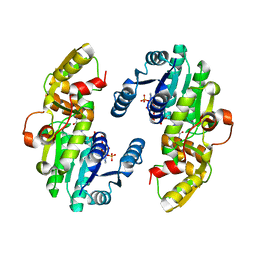 | | AtPrR1 with NADP+ | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol reductase 1 | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96212828 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7F02
 
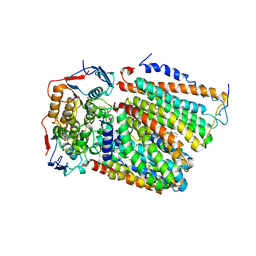 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-06-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
7F04
 
 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with Heme and ATP. | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, ADENOSINE-5'-TRIPHOSPHATE, Cytochrome c biogenesis ATP-binding export protein CcmA, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-06-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
7F03
 
 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with ANP | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-06-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
6O1S
 
 | | Structure of human plasma kallikrein protease domain with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, N-[(6-amino-2,4-dimethylpyridin-3-yl)methyl]-1-({4-[(1H-pyrazol-1-yl)methyl]phenyl}methyl)-1H-pyrazole-4-carboxamide, PHOSPHATE ION, ... | | Authors: | Partridge, J.R, Choy, R.M. | | Deposit date: | 2019-02-21 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of full-length plasma kallikrein bound to highly specific inhibitors describe a new mode of targeted inhibition.
J.Struct.Biol., 206, 2019
|
|
7VFP
 
 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with heme and single ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-09-13 | | Release date: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
7VFJ
 
 | | Cytochrome c-type biogenesis protein CcmABCD | | Descriptor: | Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, Heme exporter protein C, ... | | Authors: | Zhu, J.P, Zhang, K, Li, J, Zheng, W, Gu, M. | | Deposit date: | 2021-09-13 | | Release date: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
7X52
 
 | | Crystal structure of Bacteroides thetaiotaomicron glutamate decarboxylase BTGAD-PLP complex | | Descriptor: | ACETATE ION, Glutamate decarboxylase, MALONATE ION, ... | | Authors: | Liu, S, Du, G, Wang, Y, Wen, B, Xin, F. | | Deposit date: | 2022-03-03 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Coordinated regulation of Bacteroides thetaiotaomicron glutamate decarboxylase activity by multiple elements under different pH.
Food Chem, 403, 2023
|
|
7X4Y
 
 | | Crystal structure of Bacteroides thetaiotaomicron glutamate decarboxylase BTGAD-PLP-GABA complex | | Descriptor: | GAMMA-AMINO-BUTANOIC ACID, Glutamate decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Liu, S, Du, G, Wang, Y, Wen, B, Xin, F. | | Deposit date: | 2022-03-03 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Coordinated regulation of Bacteroides thetaiotaomicron glutamate decarboxylase activity by multiple elements under different pH.
Food Chem, 403, 2023
|
|
7X51
 
 | | Crystal structure of Bacteroides thetaiotaomicron glutamate decarboxylase BTGAD-PLP-GUA complex | | Descriptor: | GLUTARIC ACID, Glutamate decarboxylase, MALONATE ION, ... | | Authors: | Liu, S, Du, G, Wang, L, Wen, B, Xin, F. | | Deposit date: | 2022-03-03 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Coordinated regulation of Bacteroides thetaiotaomicron glutamate decarboxylase activity by multiple elements under different pH.
Food Chem, 403, 2023
|
|
7X4L
 
 | | Crystal structure of Bacteroides thetaiotaomicron glutamate decarboxylase mutant Y303F-PLP complex | | Descriptor: | Glutamate decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Liu, S, Guoming, D, Yulu, W, Boting, W, Xin, F. | | Deposit date: | 2022-03-02 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Coordinated regulation of Bacteroides thetaiotaomicron glutamate decarboxylase activity by multiple elements under different pH.
Food Chem, 403, 2023
|
|
7XVG
 
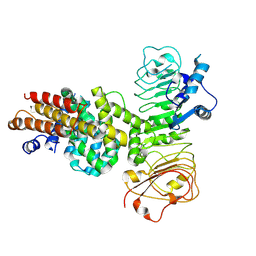 | | Cryo-EM structure of binary complex of plant NLR Sr35 and effector AvrSr35 | | Descriptor: | AvrSr35, Sr35 | | Authors: | Ouyang, S.Y, Zhao, Y.B, Li, Z.K, Liu, M.X. | | Deposit date: | 2022-05-23 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Pathogen effector AvrSr35 triggers Sr35 resistosome assembly via a direct recognition mechanism.
Sci Adv, 8, 2022
|
|
7XX2
 
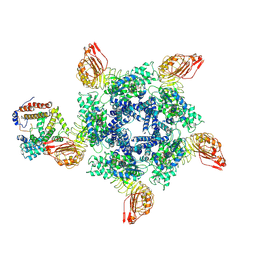 | | Cryo-EM structure of Sr35 resistosome induced by AvrSr35 R381A | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, AvrSr35, CNL9 | | Authors: | Ouyang, S.Y, Zhao, Y.B, Li, Z.K, Liu, M.X. | | Deposit date: | 2022-05-28 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Pathogen effector AvrSr35 triggers Sr35 resistosome assembly via a direct recognition mechanism.
Sci Adv, 8, 2022
|
|
7WQO
 
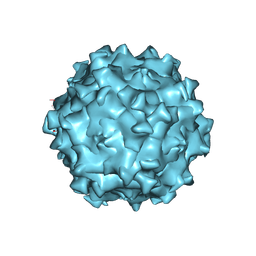 | | Structure of Adeno-associated virus serotype PHP.eB | | Descriptor: | Capsid protein VP1 | | Authors: | Xu, G, Lou, Z. | | Deposit date: | 2022-01-25 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis for the neurotropic AAV9 and the engineered AAVPHP.eB recognition with cellular receptors.
Mol Ther Methods Clin Dev, 26, 2022
|
|
7WJX
 
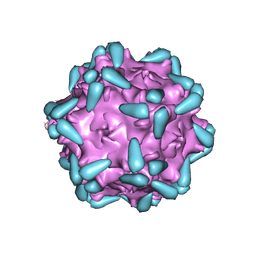 | | Adeno-associated virus serotype 9 in complex with AAVR | | Descriptor: | Capsid protein VP1, Dyslexia-associated protein KIAA0319-like protein | | Authors: | Xu, G, Lou, Z. | | Deposit date: | 2022-01-08 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis for the neurotropic AAV9 and the engineered AAVPHP.eB recognition with cellular receptors.
Mol Ther Methods Clin Dev, 26, 2022
|
|
7WJW
 
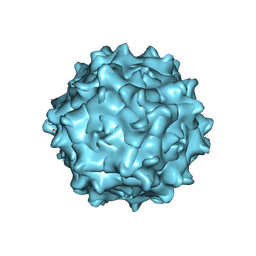 | | Structure of Adeno-associated virus serotype 9 | | Descriptor: | Capsid protein VP1 | | Authors: | Xu, G, Lou, Z. | | Deposit date: | 2022-01-08 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structural basis for the neurotropic AAV9 and the engineered AAVPHP.eB recognition with cellular receptors.
Mol Ther Methods Clin Dev, 26, 2022
|
|
7WQP
 
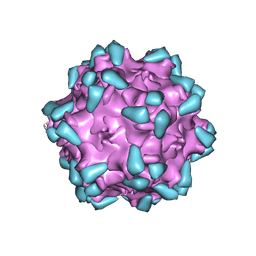 | | Adeno-associated virus serotype PHP.eB in complex with AAVR | | Descriptor: | Capsid protein VP1, Dyslexia-associated protein KIAA0319-like protein | | Authors: | Xu, G, Lou, Z. | | Deposit date: | 2022-01-25 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural basis for the neurotropic AAV9 and the engineered AAVPHP.eB recognition with cellular receptors.
Mol Ther Methods Clin Dev, 26, 2022
|
|
7XE0
 
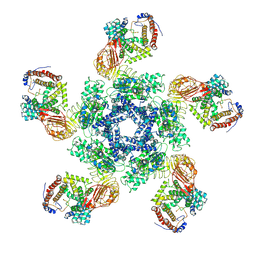 | | Cryo-EM structure of plant NLR Sr35 resistosome | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, AvrSr35, Sr35 | | Authors: | Ouyang, S.Y, Zhao, Y.B, Li, Z.K, Liu, M.X. | | Deposit date: | 2022-03-29 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Pathogen effector AvrSr35 triggers Sr35 resistosome assembly via a direct recognition mechanism.
Sci Adv, 8, 2022
|
|
7XDS
 
 | |
