1BZO
 
 | | THREE-DIMENSIONAL STRUCTURE OF PROKARYOTIC CU,ZN SUPEROXIDE DISMUTASE FROM P.LEIOGNATHI, SOLVED BY X-RAY CRYSTALLOGRAPHY. | | Descriptor: | COPPER (II) ION, PROTEIN (SUPEROXIDE DISMUTASE), URANYL (VI) ION, ... | | Authors: | Bordo, D, Matak, D, Djinovic-Carugo, K, Rosano, C, Pesce, A, Bolognesi, M, Stroppolo, M.E, Falconi, M, Battistoni, A, Desideri, A. | | Deposit date: | 1998-11-02 | | Release date: | 1999-04-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolutionary constraints for dimer formation in prokaryotic Cu,Zn superoxide dismutase.
J.Mol.Biol., 285, 1999
|
|
3ZS6
 
 | | The Structural characterization of Burkholderia pseudomallei OppA. | | Descriptor: | CHLORIDE ION, GLYCEROL, OLIGOPEPTIDE DVA, ... | | Authors: | Lassaux, P, Gourlay, L.J, Bolognesi, M. | | Deposit date: | 2011-06-23 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Structure-Based Strategy for Epitope Discovery in Burkholderia Pseudomallei Oppa Antigen.
Structure, 21, 2013
|
|
1E7Q
 
 | | GDP 4-keto-6-deoxy-D-mannose epimerase reductase S107A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYLPHOSPHATE, GDP-FUCOSE SYNTHETASE, ... | | Authors: | Rosano, C, Izzo, G, Bolognesi, M. | | Deposit date: | 2000-09-07 | | Release date: | 2000-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing the Catalytic Mechanism of Gdp-4-Keto-6-Deoxy-D-Mannose Epimerase/Reductase by Kinetic and Crystallographic Characterization of Site-Specific Mutants
J.Mol.Biol., 303, 2000
|
|
1DKE
 
 | | NI BETA HEME HUMAN HEMOGLOBIN | | Descriptor: | HEMOGLOBIN: ALPHA CHAIN, HEMOGLOBIN: BETA CHAIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Bruno, S, Bettatti, S, Mozzarelli, A, Bolognesi, M, Deriu, D, Rosano, C, Tsuneshige, A, Yonetani, T, Henry, E.R. | | Deposit date: | 1999-12-07 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Oxygen binding by alpha(Fe2+)2beta(Ni2+)2 hemoglobin crystals.
Protein Sci., 9, 2000
|
|
1E7S
 
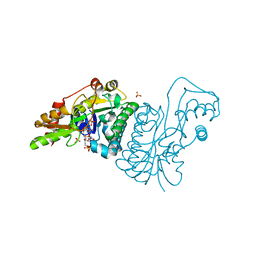 | | GDP 4-keto-6-deoxy-D-mannose epimerase reductase K140R | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYLPHOSPHATE, GDP-FUCOSE SYNTHETASE, ... | | Authors: | Rosano, C, Zuccotti, S, Izzo, G, Bolognesi, M. | | Deposit date: | 2000-09-07 | | Release date: | 2000-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the Catalytic Mechanism of Gdp-4-Keto-6-Deoxy-D-Mannose Epimerase/Reductase by Kinetic and Crystallographic Characterization of Site-Specific Mutants
J.Mol.Biol., 303, 2000
|
|
1E7R
 
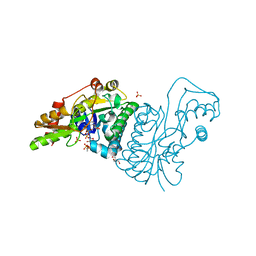 | | GDP 4-keto-6-deoxy-D-mannose epimerase reductase Y136E | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYLPHOSPHATE, GDP-FUCOSE SYNTHETASE, ... | | Authors: | Rosano, C, Izzo, G, Bolognesi, M. | | Deposit date: | 2000-09-07 | | Release date: | 2000-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing the Catalytic Mechanism of Gdp-4-Keto-6-Deoxy-D-Mannose Epimerase/Reductase by Kinetic and Crystallographic Characterization of Site-Specific Mutants
J.Mol.Biol., 303, 2000
|
|
3EKC
 
 | |
3F02
 
 | |
3F5N
 
 | |
1VHB
 
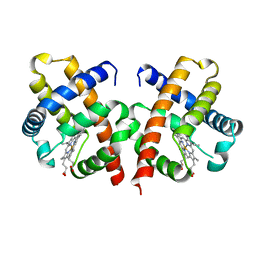 | | BACTERIAL DIMERIC HEMOGLOBIN FROM VITREOSCILLA STERCORARIA | | Descriptor: | HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tarricone, C, Galizzi, A, Coda, A, Ascenzi, P, Bolognesi, M. | | Deposit date: | 1997-02-19 | | Release date: | 1998-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Unusual structure of the oxygen-binding site in the dimeric bacterial hemoglobin from Vitreoscilla sp.
Structure, 5, 1997
|
|
1UMO
 
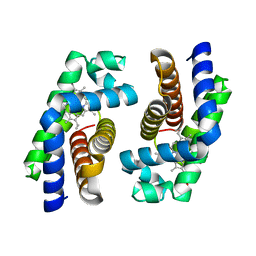 | | The crystal structure of cytoglobin: the fourth globin type discovered in man | | Descriptor: | CYTOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | de Sanctis, D, Dewilde, S, Pesce, A, Moens, L, Ascenzi, P, Hankeln, T, Burmester, T, Bolognesi, M. | | Deposit date: | 2003-08-26 | | Release date: | 2004-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of cytoglobin: the fourth globin type discovered in man displays heme hexa-coordination.
J.Mol.Biol., 336, 2004
|
|
1UX9
 
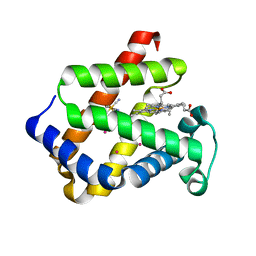 | | Mapping protein matrix cavities in human cytoglobin through Xe atom binding: a crystallographic investigation | | Descriptor: | CYTOGLOBIN, HEXACYANOFERRATE(3-), PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | De Sanctis, D, Dewilde, S, Pesce, A, Moens, L, Ascenzi, P, Hankeln, T, Burmester, T, Bolognesi, M. | | Deposit date: | 2004-02-23 | | Release date: | 2004-06-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mapping Protein Matrix Cavities in Human Cytoglobin Through Xe Atom Binding
Biochem.Biophys.Res.Commun., 316, 2004
|
|
1URV
 
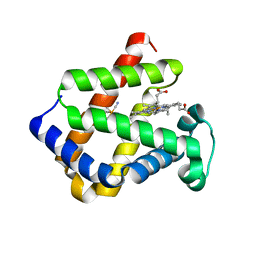 | | Crystal structure of cytoglobin: the fourth globin type discovered in man displays heme hexa-coordination | | Descriptor: | CYTOGLOBIN, HEXACYANOFERRATE(3-), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | de Sanctis, D, Dewilde, S, Pesce, A, Moens, L, Ascenzi, P, Hankeln, T, Burmester, T, Bolognesi, M. | | Deposit date: | 2003-11-11 | | Release date: | 2004-12-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Cytoglobin: The Fourth Globin Type Discovered in Man Displays Heme Hexa-Coordination.
J.Mol.Biol., 336, 2004
|
|
1UT0
 
 | | CRYSTAL STRUCTURE OF CYTOGLOBIN: THE FOURTH GLOBIN TYPE DISCOVERED IN MAN DISPLAYS HEME HEXA-COORDINATION | | Descriptor: | CYTOGLOBIN, HEXACYANOFERRATE(3-), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | De Sanctis, D, Dewilde, S, Pesce, A, Moens, L, Ascenzi, P, Hankeln, T, Burmester, T, Bolognesi, M. | | Deposit date: | 2003-12-02 | | Release date: | 2004-06-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Cytoglobin: The Fourth Globin Type Discovered in Man Displays Heme Hexa-Coordination
J.Mol.Biol., 336, 2004
|
|
1URH
 
 | | The "Rhodanese" fold and catalytic mechanism of 3-mercaptopyruvate sulfotransferases: Crystal structure of SseA from Escherichia coli | | Descriptor: | 3-MERCAPTOPYRUVATE SULFURTRANSFERASE, SULFITE ION | | Authors: | Spallarossa, A, Forlani, F, Carpen, A, Armirotti, A, Pagani, S, Bolognesi, M, Bordo, D. | | Deposit date: | 2003-10-30 | | Release date: | 2003-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The "Rhodanese" Fold and Catalytic Mechanism of 3-Mercaptopyruvate Sulfurtransferases: Crystal Structure of Ssea from Escherichia Coli
J.Mol.Biol., 335, 2004
|
|
1URY
 
 | | cytoglobin cavities | | Descriptor: | CYTOGLOBIN, HEXACYANOFERRATE(3-), PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | de Sanctis, D, Dewilde, S, Pesce, A, Moens, L, Ascenzi, P, Hankeln, T, Burmester, T, Bolognesi, M. | | Deposit date: | 2003-11-12 | | Release date: | 2004-12-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cytoglobin Cavities
Biochem.Biophys.Res.Commun., 316, 2004
|
|
1ESO
 
 | | MONOMERIC CU,ZN SUPEROXIDE DISMUTASE FROM ESCHERICHIA COLI | | Descriptor: | COPPER (II) ION, CU, ZN SUPEROXIDE DISMUTASE, ... | | Authors: | Pesce, A, Capasso, C, Battistoni, A, Folcarelli, S, Rotilio, G, Desideri, A, Bolognesi, M. | | Deposit date: | 1997-06-27 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unique structural features of the monomeric Cu,Zn superoxide dismutase from Escherichia coli, revealed by X-ray crystallography.
J.Mol.Biol., 274, 1997
|
|
8CPE
 
 | |
5FAF
 
 | | N184K pathological variant of gelsolin domain 2 (orthorhombic form) | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Boni, F, Milani, M, Ricagno, s, Bolognesi, M, de Rosa, M. | | Deposit date: | 2015-12-11 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Molecular basis of a novel renal amyloidosis due to N184K gelsolin variant.
Sci Rep, 6, 2016
|
|
7OFN
 
 | |
5M3D
 
 | | Structural tuning of CD81LEL (space group P31) | | Descriptor: | 1,2-ETHANEDIOL, CD81 antigen, PHOSPHATE ION | | Authors: | Cunha, E.S, Sfriso, P, Rojas, A.L, Roversi, P, Hospital, A, Orozco, M, Abrescia, N.G. | | Deposit date: | 2016-10-14 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Mechanism of Structural Tuning of the Hepatitis C Virus Human Cellular Receptor CD81 Large Extracellular Loop.
Structure, 25, 2017
|
|
6R85
 
 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with L-glutamate | | Descriptor: | 1,2-ETHANEDIOL, GLUTAMIC ACID, Glutamate receptor 3.3,Glutamate receptor 3.3, ... | | Authors: | Alfieri, A, Pederzoli, R, Costa, A. | | Deposit date: | 2019-03-31 | | Release date: | 2020-01-01 | | Last modified: | 2020-01-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6R8A
 
 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with L-methionine | | Descriptor: | Glutamate receptor 3.3,Glutamate receptor 3.3, METHIONINE, SODIUM ION, ... | | Authors: | Alfieri, A, Pederzoli, R, Costa, A. | | Deposit date: | 2019-04-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6R88
 
 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with glycine | | Descriptor: | CHLORIDE ION, GLYCEROL, GLYCINE, ... | | Authors: | Alfieri, A, Pederzoli, R, Costa, A. | | Deposit date: | 2019-04-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6R89
 
 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with L-cysteine | | Descriptor: | CHLORIDE ION, CYSTEINE, GLYCEROL, ... | | Authors: | Alfieri, A, Pederzoli, R, Costa, A. | | Deposit date: | 2019-04-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
