8X82
 
 | |
8X84
 
 | |
6K9N
 
 | | Rice_OTUB_like_catalytic domain | | Descriptor: | Ubiquitin thioesterase | | Authors: | Lu, L.N, Liu, L, Wang, F. | | Deposit date: | 2019-06-17 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Nat Commun, 13, 2022
|
|
8K3G
 
 | |
8TV9
 
 | | Inner Mat-T4P complex | | Descriptor: | Fimbrial protein, Maturation protein | | Authors: | Meng, R, Xing, Z, Thongchol, J, Zhang, J. | | Deposit date: | 2023-08-17 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (8.15 Å) | | Cite: | Structural basis of Acinetobacter type IV pili targeting by an RNA virus.
Nat Commun, 15, 2024
|
|
8TW2
 
 | | Acinetobacter phage AP205 T=4 VLP | | Descriptor: | Coat protein | | Authors: | Meng, R, Xing, Z, Zhang, J. | | Deposit date: | 2023-08-19 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural basis of Acinetobacter type IV pili targeting by an RNA virus.
Nat Commun, 15, 2024
|
|
8TVA
 
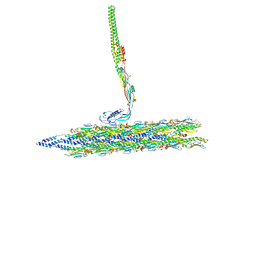 | | Outer Mat-T4P complex | | Descriptor: | Fimbrial protein, Maturation protein | | Authors: | Meng, R, Xing, Z, Thongchol, J, Zhang, J. | | Deposit date: | 2023-08-17 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (8.55 Å) | | Cite: | Structural basis of Acinetobacter type IV pili targeting by an RNA virus.
Nat Commun, 15, 2024
|
|
6KBE
 
 | | Structure of Deubiquitinase | | Descriptor: | Polyubiquitin-C, Ubiquitin thioesterase | | Authors: | Lu, L.N, Liu, L, Wang, F. | | Deposit date: | 2019-06-24 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.339 Å) | | Cite: | Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Nat Commun, 13, 2022
|
|
6K8K
 
 | | Crystal structure of Arabidopsis thaliana BIC2-CRY2 complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Cryptochrome-2, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Wang, X, Ma, L, Guan, Z, Yin, P. | | Deposit date: | 2019-06-12 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into BIC-mediated inactivation of Arabidopsis cryptochrome 2.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6IN9
 
 | | Crystal structure of MucB in complex with MucA(peri) | | Descriptor: | Sigma factor AlgU negative regulatory protein, Sigma factor AlgU regulatory protein MucB | | Authors: | Li, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis for the recognition of MucA by MucB and AlgU in Pseudomonas aeruginosa.
Febs J., 286, 2019
|
|
9BDX
 
 | | NF-kappaB RelA homo-dimer bound to CG-centric kappaB DNA | | Descriptor: | DNA (5'-D(*AP*TP*CP*AP*CP*TP*GP*GP*AP*AP*GP*TP*TP*CP*CP*CP*AP*GP*T)-3'), DNA (5'-D(P*AP*CP*TP*GP*GP*GP*AP*AP*CP*TP*TP*CP*CP*AP*GP*TP*GP*AP*T)-3'), Transcription factor p65 | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V, Ghosh, G. | | Deposit date: | 2024-04-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BDU
 
 | | NF-kappaB RelA homo-dimer bound to AT-centric kappaB DNA | | Descriptor: | DNA (5'-D(P*AP*CP*TP*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*AP*GP*TP*GP*AP*T)-3'), DNA (5'-D(P*AP*TP*CP*AP*CP*TP*GP*GP*AP*AP*TP*TP*TP*CP*CP*CP*AP*GP*T)-3'), SULFATE ION, ... | | Authors: | Biswas, T, Huang, D, Ghosh, G. | | Deposit date: | 2024-04-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BDV
 
 | | NF-kappaB RelA homo-dimer bound to TA-centric kappaB DNA | | Descriptor: | DNA (5'-D(*AP*TP*CP*AP*CP*TP*GP*GP*AP*AP*AP*TP*TP*CP*CP*CP*AP*GP*T)-3'), DNA (5'-D(P*AP*CP*TP*GP*GP*GP*AP*AP*TP*TP*TP*CP*CP*AP*GP*TP*GP*AP*T)-3'), SULFATE ION, ... | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V. | | Deposit date: | 2024-04-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BDW
 
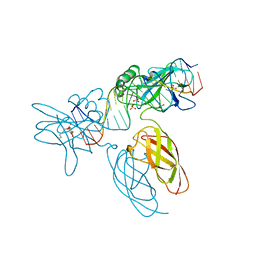 | | NF-kappaB RelA homo-dimer bound to GC-centric kappaB DNA | | Descriptor: | DNA (5'-D(P*AP*CP*TP*GP*GP*GP*AP*AP*GP*TP*TP*CP*CP*AP*GP*TP*GP*AP*T)-3'), DNA (5'-D(P*AP*TP*CP*AP*CP*TP*GP*GP*AP*AP*CP*TP*TP*CP*CP*CP*AP*GP*T)-3'), SULFATE ION, ... | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V, Huang, D, Ghosh, G. | | Deposit date: | 2024-04-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7EY5
 
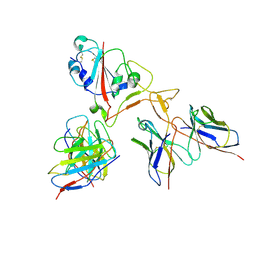 | |
7EYA
 
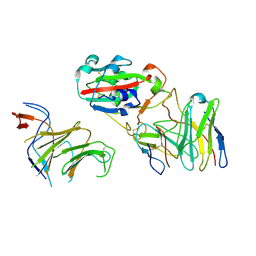 | |
7EZV
 
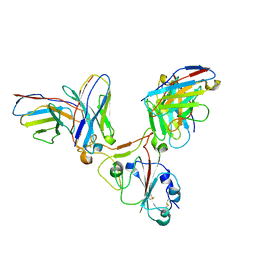 | |
7EY4
 
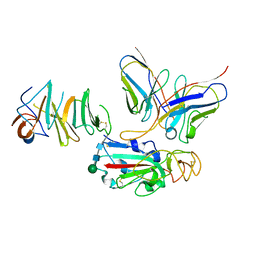 | | Local CryoEM of the SARS-CoV-2 S6PV2 in complex with BD-667 | | Descriptor: | BD-667 H, BD-667 L, Spike glycoprotein, ... | | Authors: | Liu, P.L. | | Deposit date: | 2021-05-29 | | Release date: | 2021-09-08 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Structures of SARS-CoV-2 B.1.351 neutralizing antibodies provide insights into cocktail design against concerning variants.
Cell Res., 31, 2021
|
|
7EY0
 
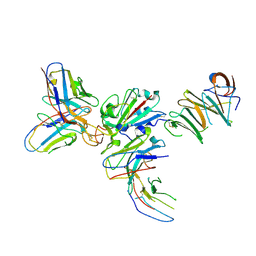 | |
6KEY
 
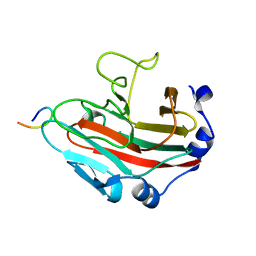 | |
6K8I
 
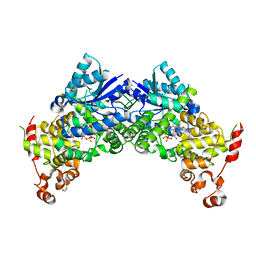 | | Crystal structure of Arabidopsis thaliana CRY2 | | Descriptor: | Cryptochrome-2, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Ma, L, Wang, X, Guan, Z, Yin, P. | | Deposit date: | 2019-06-12 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.697 Å) | | Cite: | Structural insights into BIC-mediated inactivation of Arabidopsis cryptochrome 2.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6K9P
 
 | | Structure of Deubiquitinase | | Descriptor: | Ubiquitin, Ubiquitin thioesterase | | Authors: | Lu, L.N, Liu, L, Wang, F. | | Deposit date: | 2019-06-17 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Nat Commun, 13, 2022
|
|
4HIQ
 
 | | The Structure of V122I Mutant Transthyretin in Complex with AG10 | | Descriptor: | 3-[3-(3,5-dimethyl-1H-pyrazol-4-yl)propoxy]-4-fluorobenzoic acid, Transthyretin | | Authors: | Connelly, S, Alhamadsheh, M, Graef, I, Wilson, I.A. | | Deposit date: | 2012-10-11 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | AG10 inhibits amyloidogenesis and cellular toxicity of the familial amyloid cardiomyopathy-associated V122I transthyretin.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HIS
 
 | | The Structure of V122I Mutant Transthyretin in Complex with Tafamidis | | Descriptor: | 2-(3,5-dichlorophenyl)-1,3-benzoxazole-6-carboxylic acid, Transthyretin | | Authors: | Connelly, S, Alhamadsheh, M, Graef, I, Wilson, I.A. | | Deposit date: | 2012-10-11 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | AG10 inhibits amyloidogenesis and cellular toxicity of the familial amyloid cardiomyopathy-associated V122I transthyretin.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6KPC
 
 | | Crystal structure of an agonist bound GPCR | | Descriptor: | 7-[(6aR,9R,10aR)-1-Hydroxy-9-(hydroxymethyl)-6,6-dimethyl-6a,7,8,9,10,10a-hexahydro-6H-benzo[c]chromen-3-yl]- 7-methyloctanenitrile, Cannabinoid receptor 2,Endolysin | | Authors: | Li, X.T, Hua, T, Wu, L.J, Makriyannis, A, Wu, M, Liu, Z.J. | | Deposit date: | 2019-08-15 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-GiComplex Structures.
Cell, 180, 2020
|
|
