6YE1
 
 | | Human Ecto-5'-nucleotidase (CD73) in complex with the AMPCP derivative A894 (compound 2n in publication) in the closed form (crystal form IV) | | Descriptor: | 5'-nucleotidase, ZINC ION, [(2~{R},3~{S},4~{R},5~{R})-5-[2-chloranyl-6-(cyclopentylamino)purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxymethylphosphonic acid | | Authors: | Scaletti, E, Strater, N. | | Deposit date: | 2020-03-23 | | Release date: | 2021-01-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Discovery of Potent and Selective Methylenephosphonic Acid CD73 Inhibitors.
J.Med.Chem., 64, 2021
|
|
6JZZ
 
 | | The crystal structure of AAR-C294S in complex with ADO. | | Descriptor: | Aldehyde decarbonylase, FE (II) ION, HEXADECAN-1-OL, ... | | Authors: | Zhang, H.M, Li, M, Gao, Y. | | Deposit date: | 2019-05-04 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | Structural insights into catalytic mechanism and product delivery of cyanobacterial acyl-acyl carrier protein reductase.
Nat Commun, 11, 2020
|
|
6JZY
 
 | | Crystal structure of AAR with NADPH and stearyl in complex with ADO binding a long chain carbohydrate | | Descriptor: | Aldehyde decarbonylase, FE (II) ION, HEXADECAN-1-OL, ... | | Authors: | Zhang, H.M, Li, M, Gao, Y. | | Deposit date: | 2019-05-04 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into catalytic mechanism and product delivery of cyanobacterial acyl-acyl carrier protein reductase.
Nat Commun, 11, 2020
|
|
2OJU
 
 | | X-ray structure of complex of human cyclophilin J with cyclosporin A | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE-LIKE 3 | | Authors: | Xia, Z, Huang, L. | | Deposit date: | 2007-01-14 | | Release date: | 2008-01-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting Cyclophilin J, a Novel Peptidyl-Prolyl Isomerase, Can Induce Cellular G1/S Arrest and Repress the Growth of Hepatocellular Carcinoma
To be Published
|
|
6YE2
 
 | |
2OK3
 
 | | X-ray structure of human cyclophilin J at 2.0 angstrom | | Descriptor: | NICKEL (II) ION, Peptidyl-prolyl cis-trans isomerase-like 3 | | Authors: | Xia, Z. | | Deposit date: | 2007-01-15 | | Release date: | 2008-01-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeting Cyclophilin J, a novel peptidyl-prolyl isomerase, can induce cellular G1/S arrest and repress the growth of Hepatocellular carcinoma
To be Published
|
|
6JZQ
 
 | |
6JZU
 
 | | The crystal structure of acyl-acyl carrier protein (acyl-ACP) reductase (AAR) in complex with aldehyde deformylating oxygenase (ADO) | | Descriptor: | Aldehyde decarbonylase, FE (II) ION, HEXADECAN-1-OL, ... | | Authors: | Zhang, H.M, Li, M, Gao, Y. | | Deposit date: | 2019-05-03 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.181 Å) | | Cite: | Structural insights into catalytic mechanism and product delivery of cyanobacterial acyl-acyl carrier protein reductase.
Nat Commun, 11, 2020
|
|
6IH9
 
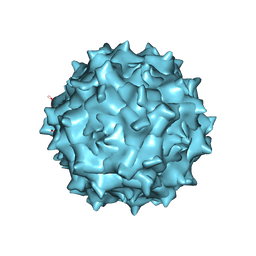 | | Adeno-Associated Virus 2 at 2.8 ang | | Descriptor: | Capsid protein VP1 | | Authors: | Lou, Z.Y, Zhang, R. | | Deposit date: | 2018-09-29 | | Release date: | 2019-07-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Adeno-associated virus 2 bound to its cellular receptor AAVR.
Nat Microbiol, 4, 2019
|
|
7YJ3
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with human ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-07-19 | | Release date: | 2023-07-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
7YV8
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with golden hamster ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike glycoprotein, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Chai, Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-08-18 | | Release date: | 2023-07-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
7YVU
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with mouse ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Chai, Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-08-19 | | Release date: | 2023-07-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
1XMZ
 
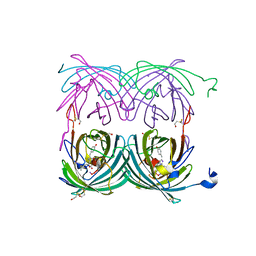 | | Crystal structure of the dark state of kindling fluorescent protein kfp from anemonia sulcata | | Descriptor: | BETA-MERCAPTOETHANOL, GFP-like non-fluorescent chromoprotein FP595 chain 1, GFP-like non-fluorescent chromoprotein FP595 chain 2 | | Authors: | Quillin, M.L, Anstrom, D.M, Shu, X, O'Leary, S, Kallio, K, Chudakov, D.M, Remington, S.J. | | Deposit date: | 2004-10-04 | | Release date: | 2005-04-19 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Kindling Fluorescent Protein from Anemonia sulcata: Dark-State Structure at 1.38 Resolution
Biochemistry, 44, 2005
|
|
9KCH
 
 | |
9K49
 
 | |
8GRY
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with rat ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Chai, Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-09-03 | | Release date: | 2023-07-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
8H06
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.4/5 RBD in complex with human ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-09-28 | | Release date: | 2023-07-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
8H5C
 
 | | Structure of SARS-CoV-2 Omicron BA.2.75 RBD in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Z.N, Bai, B, Liu, K.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-10-12 | | Release date: | 2023-07-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
6LA3
 
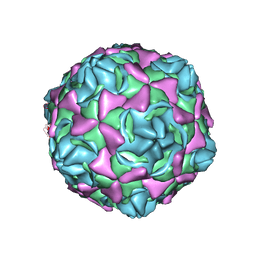 | | Cryo-EM structure of full echovirus 11 particle at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2019-11-11 | | Release date: | 2020-10-07 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | Molecular and structural basis of Echovirus 11 infection by using the dual-receptor system of CD55 and FcRn.
Chin.Sci.Bull., 2020
|
|
6LAO
 
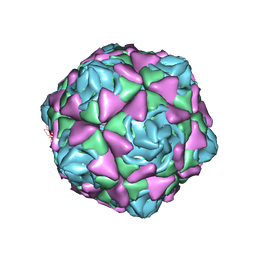 | |
6LBO
 
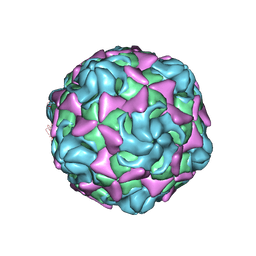 | | Cryo-EM structure of echovirus 11 empty particle at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2019-11-14 | | Release date: | 2020-10-07 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Molecular and structural basis of Echovirus 11 infection by using the dual-receptor system of CD55 and FcRn.
Chin.Sci.Bull., 2020
|
|
6LBQ
 
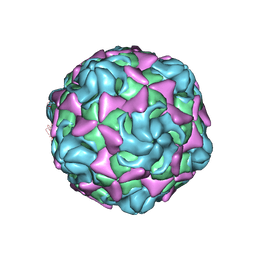 | | Cryo-EM structure of echovirus 11 empty particle at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2019-11-14 | | Release date: | 2020-10-07 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular and structural basis of Echovirus 11 infection by using the dual-receptor system of CD55 and FcRn.
Chin.Sci.Bull., 2020
|
|
6LIX
 
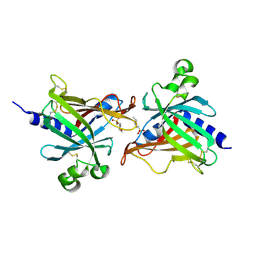 | | CRL Protein of Arabidopsis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Chromophore lyase CRL, chloroplastic | | Authors: | Wang, F.F, Guan, K.L, Sun, P.K, Xing, W.M. | | Deposit date: | 2019-12-13 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.385 Å) | | Cite: | The Arabidopsis CRUMPLED LEAF protein, a homolog of the cyanobacterial bilin lyase, retains the bilin-binding pocket for a yet unknown function.
Plant J., 104, 2020
|
|
6LA4
 
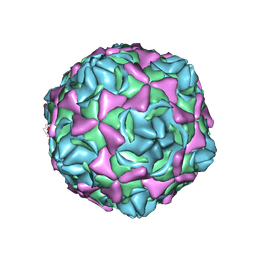 | | Cryo-EM structure of full echovirus 11 particle at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2019-11-11 | | Release date: | 2020-10-07 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | Molecular and structural basis of Echovirus 11 infection by using the dual-receptor system of CD55 and FcRn.
Chin.Sci.Bull., 2020
|
|
6L9J
 
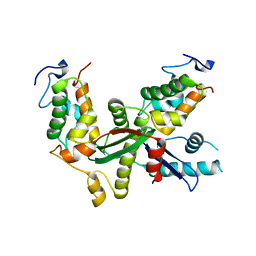 | | Structure of yeast Snf5 and Swi3 subcomplex | | Descriptor: | GLYCEROL, SWI/SNF chromatin-remodeling complex subunit SNF5, SWI/SNF complex subunit SWI3 | | Authors: | Long, J, Zhou, H. | | Deposit date: | 2019-11-10 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.642 Å) | | Cite: | Snf5 and Swi3 subcomplex formation is required for SWI/SNF complex function in yeast.
Biochem.Biophys.Res.Commun., 526, 2020
|
|
