5X3I
 
 | | Kfla1895 D451A mutant | | Descriptor: | GLYCEROL, Glycoside hydrolase family 31, SULFATE ION | | Authors: | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | Deposit date: | 2017-02-06 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Glycoside hydrolase mutant
To Be Published
|
|
5X6C
 
 | | Crystal structure of SepRS-SepCysE from Methanocaldococcus jannaschii | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, O-phosphoserine--tRNA(Cys) ligase, SULFATE ION, ... | | Authors: | Chen, M, Kato, K, Yao, M. | | Deposit date: | 2017-02-21 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Structural basis for tRNA-dependent cysteine biosynthesis
Nat Commun, 8, 2017
|
|
5X6B
 
 | |
5X32
 
 | | Crystal structure of D-mannose isomerase | | Descriptor: | N-acylglucosamine 2-epimerase, PHOSPHATE ION | | Authors: | Kato, K, Saburi, W, Yao, M. | | Deposit date: | 2017-02-03 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Biochemical and structural characterization of Marinomonas mediterranead-mannose isomerase Marme_2490 phylogenetically distant from known enzymes
Biochimie, 144, 2018
|
|
5ZCC
 
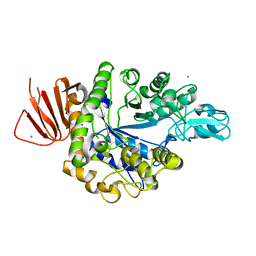 | | Crystal structure of Alpha-glucosidase in complex with maltose | | Descriptor: | Alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kato, K, Saburi, W, Yao, M. | | Deposit date: | 2018-02-16 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Function and structure of GH13_31 alpha-glucosidase with high alpha-(1→4)-glucosidic linkage specificity and transglucosylation activity.
FEBS Lett., 592, 2018
|
|
5ZCE
 
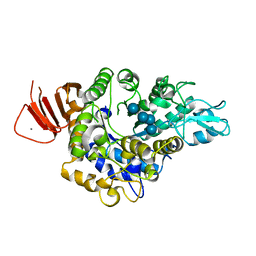 | | Crystal structure of Alpha-glucosidase in complex with maltotetraose | | Descriptor: | Alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kato, K, Saburi, W, Yao, M. | | Deposit date: | 2018-02-16 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.555 Å) | | Cite: | Function and structure of GH13_31 alpha-glucosidase with high alpha-(1→4)-glucosidic linkage specificity and transglucosylation activity.
FEBS Lett., 592, 2018
|
|
5XW7
 
 | |
5ZCD
 
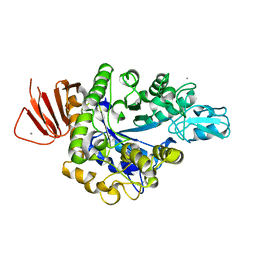 | | Crystal structure of Alpha-glucosidase in complex with maltotriose | | Descriptor: | Alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kato, K, Saburi, W, Yao, M. | | Deposit date: | 2018-02-16 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | Function and structure of GH13_31 alpha-glucosidase with high alpha-(1→4)-glucosidic linkage specificity and transglucosylation activity.
FEBS Lett., 592, 2018
|
|
5ZCB
 
 | | Crystal structure of Alpha-glucosidase | | Descriptor: | Alpha-glucosidase, CALCIUM ION | | Authors: | Kato, K, Saburi, W, Yao, M. | | Deposit date: | 2018-02-16 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Function and structure of GH13_31 alpha-glucosidase with high alpha-(1→4)-glucosidic linkage specificity and transglucosylation activity.
FEBS Lett., 592, 2018
|
|
8H25
 
 | | Lacticaseibacillus casei GH35 beta-galactosidase LBCZ_0230 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-galactosidase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Saburi, W, Ota, T, Kato, K, Tagami, T, Yamashita, K, Yao, M, Mori, H. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Function and Structure of Lacticaseibacillus casei GH35 beta-Galactosidase LBCZ_0230 with High Hydrolytic Activity to Lacto- N -biose I and Galacto- N -biose.
J Appl Glycosci (1999), 70, 2023
|
|
8IMZ
 
 | | Cryo-EM structure of mouse Piezo1-MDFIC complex (composite map) | | Descriptor: | MyoD family inhibitor domain-containing protein, Piezo-type mechanosensitive ion channel component 1 | | Authors: | Zhou, Z, Ma, X, Lin, Y, Cheng, D, Bavi, N, Li, J.V, Sutton, D, Yao, M, Harvey, N, Corry, B, Zhang, Y, Cox, C.D. | | Deposit date: | 2023-03-07 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | MyoD-family inhibitor proteins act as auxiliary subunits of Piezo channels.
Science, 381, 2023
|
|
8IDS
 
 | | Crystal structure of Bacillus sp. AHU2216 GH13_31 Alpha-glucosidase E256Q/N258P in complex with maltotriose | | Descriptor: | Alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Auiewiriyanukul, W, Saburi, W, Yu, J, Kato, K, Yao, M, Mori, H. | | Deposit date: | 2023-02-14 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Alteration of Substrate Specificity and Transglucosylation Activity of GH13_31 alpha-Glucosidase from Bacillus sp. AHU2216 through Site-Directed Mutagenesis of Asn258 on beta → alpha Loop 5.
Molecules, 28, 2023
|
|
8IBK
 
 | | Crystal structure of Bacillus sp. AHU2216 GH13_31 Alpha-glucosidase E256Q/N258G in complex with maltotriose | | Descriptor: | Alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Auiewiriyanukul, W, Saburi, W, Yu, J, Kato, K, Yao, M, Mori, H. | | Deposit date: | 2023-02-10 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Alteration of Substrate Specificity and Transglucosylation Activity of GH13_31 alpha-Glucosidase from Bacillus sp. AHU2216 through Site-Directed Mutagenesis of Asn258 on beta → alpha Loop 5.
Molecules, 28, 2023
|
|
8HRH
 
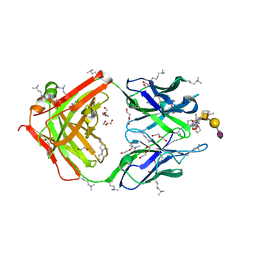 | | SN-131/1B2 anti-MUC1 antibody with a glycopeptide | | Descriptor: | 1-ACETYL-L-PROLINE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALANINE, ... | | Authors: | Wakui, H, Horidome, C, Yao, M, Ose, T, Nishimura, S.-I. | | Deposit date: | 2022-12-15 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural and molecular insight into antibody recognition of dynamic neoepitopes in membrane tethered MUC1 of pancreatic cancer cells and secreted exosomes.
Rsc Chem Biol, 4, 2023
|
|
7CMC
 
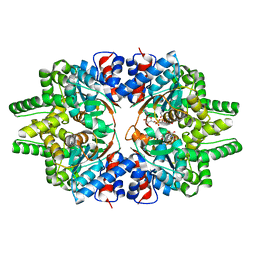 | | CRYSTAL STRUCTURE OF DEOXYHYPUSINE SYNTHASE FROM PYROCOCCUS HORIKOSHII | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Probable deoxyhypusine synthase | | Authors: | Yu, J, Gai, Z.Q, Okada, C, Yao, M. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Flexible NAD+Binding in Deoxyhypusine Synthase Reflects the Dynamic Hypusine Modification of Translation Factor IF5A.
Int J Mol Sci, 21, 2020
|
|
3WSU
 
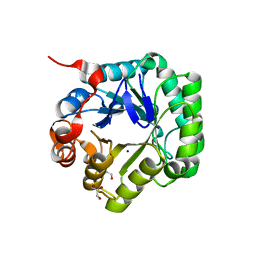 | | Crystal structure of beta-mannanase from Streptomyces thermolilacinus | | Descriptor: | Beta-mannanase, GLYCEROL, SODIUM ION | | Authors: | Kumagai, Y, Yamashita, K, Okuyama, M, Hatanaka, T, Yao, M, Kimura, A. | | Deposit date: | 2014-03-26 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The loop structure of Actinomycete glycoside hydrolase family 5 mannanases governs substrate recognition
Febs J., 282, 2015
|
|
3W9Z
 
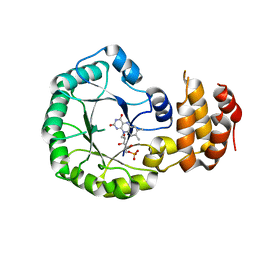 | | Crystal structure of DusC | | Descriptor: | FLAVIN MONONUCLEOTIDE, tRNA-dihydrouridine synthase C | | Authors: | Chen, M, Yu, J, Tanaka, Y, Tanaka, I, Yao, M. | | Deposit date: | 2013-04-19 | | Release date: | 2013-07-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of dihydrouridine synthase C (DusC) from Escherichia coli
Acta Crystallogr.,Sect.F, 69, 2013
|
|
7E9X
 
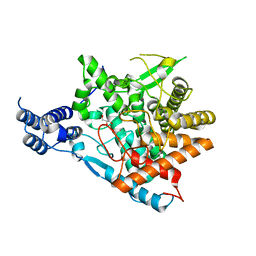 | | Trehalase of Arabidopsis thaliana acid mutant -D380A | | Descriptor: | GLYCEROL, Trehalase | | Authors: | Taguchi, Y, Saburi, W, Yu, J, Imai, R, Yao, M, Mori, H. | | Deposit date: | 2021-03-05 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | pH-dependent alteration of substrate specificity of plant trehalase and its molecular mechanism
To Be Published
|
|
7E9U
 
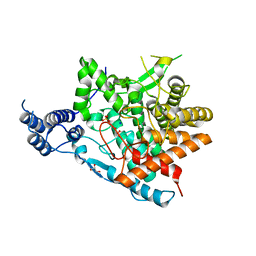 | | Trehalase of Arabidopsis thaliana | | Descriptor: | GLYCEROL, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Taguchi, Y, Saburi, W, Yu, J, Imai, R, Yao, M, Mori, H. | | Deposit date: | 2021-03-05 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | pH-dependent alteration of substrate specificity of plant trehalase and its molecular mechanism
To Be Published
|
|
7EAW
 
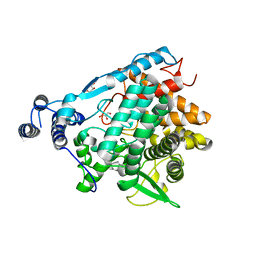 | | Trehalase of Arabidopsis thaliana acid mutant -D380A trehalose complex | | Descriptor: | GLYCEROL, Trehalase, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Taguchi, Y, Saburi, W, Yu, J, Imai, R, Yao, M, Mori, H. | | Deposit date: | 2021-03-08 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | pH-dependent alteration of substrate specificity of plant trehalase and its molecular mechanism
To Be Published
|
|
7F82
 
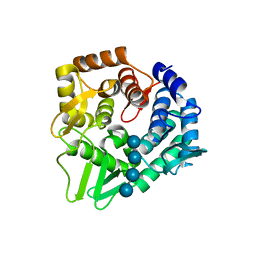 | | Structure of the bacterial cellulose synthase subunit Z in complex with cellooligosaccharides from Enterobacter sp. CJF-002 | | Descriptor: | Glucanase, S,R MESO-TARTARIC ACID, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Fujiwara, T, Fujishima, A, Yao, M. | | Deposit date: | 2021-06-30 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural snapshot of a glycoside hydrolase family 8 endo-beta-1,4-glucanase capturing the state after cleavage of the scissile bond.
Acta Crystallogr.,Sect.D, 78, 2022
|
|
7F81
 
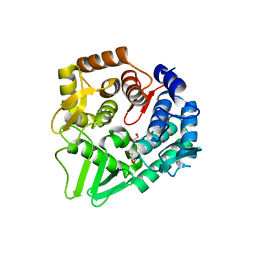 | | Structure of the bacterial cellulose synthase subunit Z from Enterobacter sp. CJF-002 | | Descriptor: | GLYCEROL, Glucanase, S,R MESO-TARTARIC ACID | | Authors: | Fujiwara, T, Fujishima, A, Yao, M. | | Deposit date: | 2021-06-30 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural snapshot of a glycoside hydrolase family 8 endo-beta-1,4-glucanase capturing the state after cleavage of the scissile bond.
Acta Crystallogr.,Sect.D, 78, 2022
|
|
7F3A
 
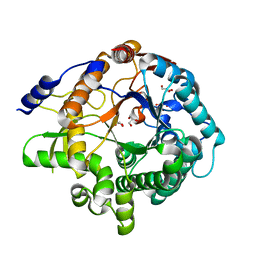 | | Arabidopsis thaliana GH1 beta-glucosidase AtBGlu42 | | Descriptor: | Beta-glucosidase 42, GLYCEROL | | Authors: | Horikoshi, S, Saburi, W, Yu, J, Yao, M. | | Deposit date: | 2021-06-16 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate specificity of glycoside hydrolase family 1 beta-glucosidase AtBGlu42 from Arabidopsis thaliana and its molecular mechanism.
Biosci.Biotechnol.Biochem., 86, 2022
|
|
3W37
 
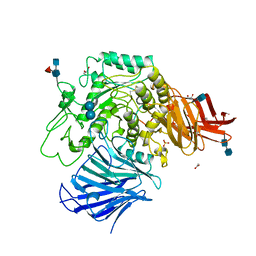 | | Sugar beet alpha-glucosidase with acarbose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Tagami, T, Yamashita, K, Okuyama, M, Mori, H, Yao, M, Kimura, A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-05-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis for the recognition of long-chain substrates by plant & alpha-glucosidase
J.Biol.Chem., 288, 2013
|
|
3W38
 
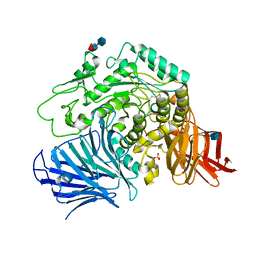 | | Sugar beet alpha-glucosidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-glucosidase, SULFATE ION, ... | | Authors: | Tagami, T, Yamashita, K, Okuyama, M, Mori, H, Yao, M, Kimura, A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-05-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Molecular basis for the recognition of long-chain substrates by plant & alpha-glucosidase
J.Biol.Chem., 288, 2013
|
|
