3C8G
 
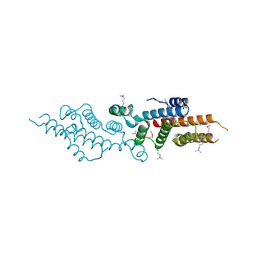 | | Crystal structure of a possible transciptional regulator YggD from Shigella flexneri 2a str. 2457T | | Descriptor: | ACETATE ION, Putative transcriptional regulator | | Authors: | Tan, K, Borovilos, M, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-12 | | Release date: | 2008-02-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The mannitol operon repressor MtlR belongs to a new class of transcription regulators in bacteria.
J.Biol.Chem., 284, 2009
|
|
3BP1
 
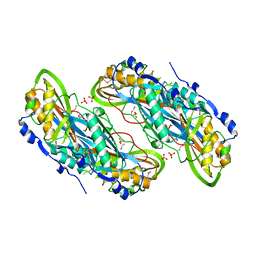 | | Crystal structure of putative 7-cyano-7-deazaguanine reductase QueF from Vibrio cholerae O1 biovar eltor | | Descriptor: | GUANINE, MAGNESIUM ION, NADPH-dependent 7-cyano-7-deazaguanine reductase, ... | | Authors: | Kim, Y, Zhou, M, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-18 | | Release date: | 2008-01-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | High-resolution structure of the nitrile reductase QueF combined with molecular simulations provide insight into enzyme mechanism.
J.Mol.Biol., 404, 2010
|
|
3BWG
 
 | | The crystal structure of possible transcriptional regulator YydK from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | 1,2-ETHANEDIOL, Uncharacterized HTH-type transcriptional regulator yydK | | Authors: | Tan, K, Zhou, M, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-09 | | Release date: | 2008-01-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The crystal structure of possible transcriptional regulator YydK from Bacillus subtilis subsp. subtilis str. 168.
To be Published
|
|
3BY6
 
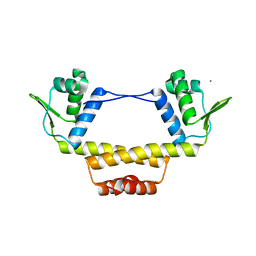 | |
3C87
 
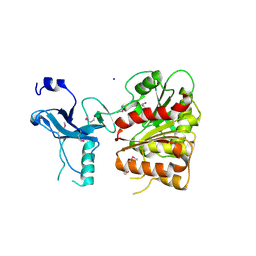 | | Crystal structure of the enterobactin esterase FES from Shigella flexneri in the presence of enterobactin | | Descriptor: | Enterochelin esterase, PHOSPHATE ION, SODIUM ION | | Authors: | Kim, Y, Maltseva, N, Abergel, R, Raymond, K, Holzle, D, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-11 | | Release date: | 2008-02-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Siderophore Mediated Iron Acquisition: Structure and Specificity of Enterobactin Esterase from Shigella flexneri.
To be Published
|
|
3C9P
 
 | | Crystal structure of uncharacterized protein SP1917 | | Descriptor: | 1,2-ETHANEDIOL, Uncharacterized protein SP1917 | | Authors: | Chang, C, Zhou, M, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-18 | | Release date: | 2008-02-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of uncharacterized protein SP1917.
To be Published
|
|
3C8I
 
 | |
3C8H
 
 | | Crystal structure of the enterobactin esterase FES from Shigella flexneri in the presence of 2,3-Di-hydroxy-N-benzoyl-serine | | Descriptor: | Enterochelin esterase | | Authors: | Kim, Y, Maltseva, N, Abergel, R, Holzle, D, Raymond, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-12 | | Release date: | 2008-02-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Siderophore Mediated Iron Acquisition: Structure and Specificity of Enterobactin Esterase from Shigella flexneri.
To be Published
|
|
3BWL
 
 | | Crystal structure of PAS domain of HTR-like protein from Haloarcula marismortui | | Descriptor: | 1H-INDOLE-3-CARBALDEHYDE, MAGNESIUM ION, Sensor protein | | Authors: | Osipiuk, J, Zhou, M, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-09 | | Release date: | 2008-01-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | X-ray crystal structure of PAS domain of HTR-like protein from Haloarcula marismortui.
To be Published
|
|
3BUU
 
 | |
1RZ2
 
 | | 1.6A crystal structure of the protein BA4783/Q81L49 (similar to sortase B) from Bacillus anthracis. | | Descriptor: | conserved hypothetical protein BA4783 | | Authors: | Wu, R, Zhang, R, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-12-23 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of sortase B from Staphylococcus aureus and Bacillus anthracis reveal catalytic amino acid triad in the active site.
Structure, 12, 2004
|
|
4WRP
 
 | | The C-terminal domain of gene product lpg0944 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | Uncharacterized protein | | Authors: | Cuff, M.E, Evdokimova, E, Egorova, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-10-24 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The C-terminal domain of gene product lpg0944 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1.
To Be Published
|
|
5E2E
 
 | | Crystal Structure of Beta-lactamase Precursor BlaA from Yersinia enterocolitica | | Descriptor: | Beta-lactamase | | Authors: | Kim, Y, Joachimiak, G, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-10-01 | | Release date: | 2015-10-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Beta-lactamase Precursor BlaA from Yersinia enterocolitica
To Be Published
|
|
6CP8
 
 | | Contact-dependent growth inhibition toxin-immunity protein complex from from E. coli 3006 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CdiA, CdiI, ... | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Convergent Evolution of the Barnase/EndoU/Colicin/RelE (BECR) Fold in Antibacterial tRNase Toxins.
Structure, 27, 2019
|
|
6CP9
 
 | | Contact-dependent growth inhibition toxin - immunity protein complex from Klebsiella pneumoniae 342 | | Descriptor: | CdiA, CdiI | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Convergent Evolution of the Barnase/EndoU/Colicin/RelE (BECR) Fold in Antibacterial tRNase Toxins.
Structure, 27, 2019
|
|
5E2F
 
 | | Crystal Structure of Beta-lactamase class D from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase YbxI, CALCIUM ION | | Authors: | Kim, Y, Joachimiak, G, Endres, M, Babnigg, G, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-10-01 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Beta-lactamase class D from Bacillus subtilis
To Be Published
|
|
5E2G
 
 | | Crystal Structure of D-alanine Carboxypeptidase AmpC from Burkholderia cenocepacia | | Descriptor: | ACETIC ACID, Beta-lactamase, THIOCYANATE ION | | Authors: | Kim, Y, Joachimiak, G, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-10-01 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Crystal Structure of D-alanine Carboxypeptidase AmpC from Burkholderia cenocepacia
To Be Published
|
|
5E3E
 
 | | Crystal structure of CdiA-CT/CdiI complex from Y. kristensenii 33638 | | Descriptor: | CdiI immunity protein, Large exoprotein involved in heme utilization or adhesion, SODIUM ION | | Authors: | Michalska, K, Joachimiak, G, Jedrzejczak, R, Goulding, C.W, Joachimiak, A, Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-10-02 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The CDI toxin of Yersinia kristensenii is a novel bacterial member of the RNase A superfamily.
Nucleic Acids Res., 45, 2017
|
|
1NG5
 
 | |
5C0P
 
 | | The crystal structure of endo-arabinase from Bacteroides thetaiotaomicron VPI-5482 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Endo-arabinase, ... | | Authors: | Tan, K, Cuff, M, Joachimiak, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-06-12 | | Release date: | 2015-07-01 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | The crystal structure of endo-arabinase from Bacteroides thetaiotaomicron VPI-5482
To Be Published
|
|
1P99
 
 | | 1.7A crystal structure of protein PG110 from Staphylococcus aureus | | Descriptor: | GLYCINE, Hypothetical protein PG110, METHIONINE | | Authors: | Zhang, R, Zhou, M, Joachimiak, G, Schneewind, O, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-05-09 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The membrane-associated lipoprotein-9 GmpC from Staphylococcus aureus binds the dipeptide GlyMet via side chain interactions.
Biochemistry, 43, 2004
|
|
6U13
 
 | | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the hydrolyzed antibiotic moxalactam | | Descriptor: | (2R)-2-((R)-CARBOXY{[CARBOXY(4-HYDROXYPHENYL)ACETYL]AMINO}METHOXYMETHYL)-5-METHYLENE-5,6-DIHYDRO-2H-1,3-OXAZINE-4-CARBO XYLIC ACID, 1,2-ETHANEDIOL, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-15 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the hydrolyzed antibiotic moxalactam.
To Be Published
|
|
6U2Y
 
 | | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the hydrolyzed moxalactam and two Ni ions | | Descriptor: | (2R)-2-((R)-CARBOXY{[CARBOXY(4-HYDROXYPHENYL)ACETYL]AMINO}METHOXYMETHYL)-5-METHYLENE-5,6-DIHYDRO-2H-1,3-OXAZINE-4-CARBO XYLIC ACID, 1,2-ETHANEDIOL, NICKEL (II) ION, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-20 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the hydrolyzed moxalactam and two Ni ions
To Be Published
|
|
6U10
 
 | | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the inhibitor captopril | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, L-CAPTOPRIL, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-15 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the inhibitor captopril.
To Be Published
|
|
6UAG
 
 | |
