3ZEE
 
 | | Electron cyro-microscopy helical reconstruction of Par-3 N terminal domain | | 分子名称: | PARTITIONING DEFECTIVE 3 HOMOLOG | | 著者 | Zhang, Y, Wang, W, Chen, J, Zhang, K, Gao, F, Gong, W, Zhang, M, Sun, F, Feng, W. | | 登録日 | 2012-12-05 | | 公開日 | 2013-10-16 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (6.1 Å) | | 主引用文献 | Structural Insights Into the Intrinsic Self-Assembly of Par-3 N-Terminal Domain.
Structure, 21, 2013
|
|
1THD
 
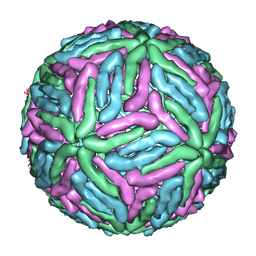 | | COMPLEX ORGANIZATION OF DENGUE VIRUS E PROTEIN AS REVEALED BY 9.5 ANGSTROM CRYO-EM RECONSTRUCTION | | 分子名称: | Major envelope protein E | | 著者 | Zhang, Y, Zhang, W, Ogata, S, Clements, D, Strauss, J.H, Baker, T.S, Kuhn, R.J, Rossmann, M.G. | | 登録日 | 2004-06-01 | | 公開日 | 2004-09-28 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (9.5 Å) | | 主引用文献 | Conformational changes of the flavivirus e glycoprotein
Structure, 12, 2004
|
|
1TGE
 
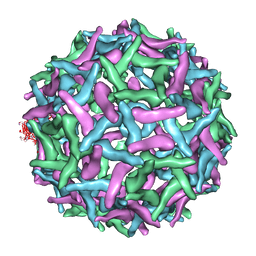 | | The structure of immature Dengue virus at 12.5 angstrom | | 分子名称: | envelope glycoprotein | | 著者 | Zhang, Y, Zhang, W, Ogata, S, Clements, D, Strauss, J.H, Baker, T.S, Kuhn, R.J, Rossmann, M.G. | | 登録日 | 2004-05-28 | | 公開日 | 2004-09-28 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (12.5 Å) | | 主引用文献 | Conformational changes of the flavivirus e glycoprotein.
Structure, 12, 2004
|
|
1TG8
 
 | | The structure of Dengue virus E glycoprotein | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, envelope glycoprotein | | 著者 | Zhang, Y, Zhang, W, Ogata, S, Clements, D, Strauss, J.H, Baker, T.S, Rossmann, M.G. | | 登録日 | 2004-05-28 | | 公開日 | 2004-09-28 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Conformational changes of the flavivirus e glycoprotein
Structure, 12, 2004
|
|
8YM1
 
 | |
5VAI
 
 | | Cryo-EM structure of the activated Glucagon-like peptide-1 receptor in complex with G protein | | 分子名称: | Glucagon-like peptide 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Zhang, Y, Sun, B, Feng, D, Hu, H, Chu, M, Qu, Q, Tarrasch, J.T, Li, S, Kobilka, T.S, Kobilka, B.K, Skiniotis, G. | | 登録日 | 2017-03-27 | | 公開日 | 2017-05-24 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein.
Nature, 546, 2017
|
|
5V22
 
 | |
1T8W
 
 | |
1T8S
 
 | |
1T8Y
 
 | |
1T8R
 
 | |
6J1L
 
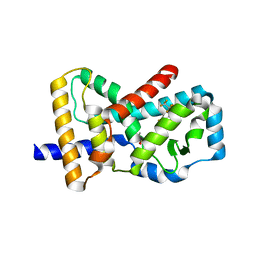 | | Crystal Structure Analysis of the ROR gamma(C455E) | | 分子名称: | 2-[4-(ethylsulfonyl)phenyl]-N-[2'-fluoro-4'-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)[1,1'-biphenyl]-4-yl]acetamide, Nuclear receptor ROR-gamma | | 著者 | zhang, Y, Li, C.C, wu, X.S. | | 登録日 | 2018-12-28 | | 公開日 | 2019-05-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Discovery and Characterization of XY101, a Potent, Selective, and Orally Bioavailable ROR gamma Inverse Agonist for Treatment of Castration-Resistant Prostate Cancer.
J.Med.Chem., 62, 2019
|
|
6VRJ
 
 | |
6VYM
 
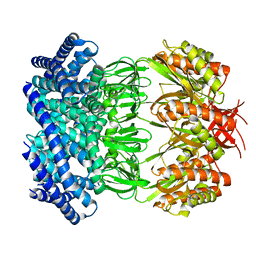 | | Cryo-EM structure of mechanosensitive channel MscS in PC-18:1 nanodiscs treated with beta-cyclodextran | | 分子名称: | Mechanosensitive channel MscS | | 著者 | Zhang, Y, Daday, C, Gu, R, Cox, C.D, Martinac, B, Groot, B, Walz, T. | | 登録日 | 2020-02-27 | | 公開日 | 2021-02-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Visualization of the mechanosensitive ion channel MscS under membrane tension.
Nature, 590, 2021
|
|
6VYK
 
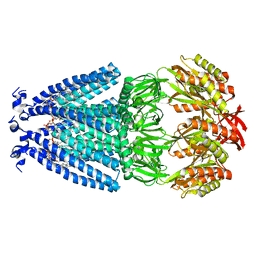 | | Cryo-EM structure of mechanosensitive channel MscS in PC-18:1 nanodiscs | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Mechanosensitive channel MscS | | 著者 | Zhang, Y, Daday, C, Gu, R, Cox, C.D, Martinac, B, Groot, B, Walz, T. | | 登録日 | 2020-02-27 | | 公開日 | 2021-02-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Visualization of the mechanosensitive ion channel MscS under membrane tension.
Nature, 590, 2021
|
|
6VYL
 
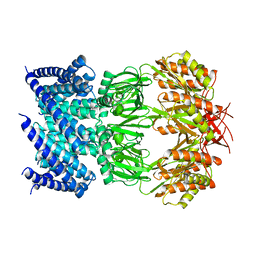 | | Cryo-EM structure of mechanosensitive channel MscS in PC-10 nanodiscs | | 分子名称: | Mechanosensitive channel MscS | | 著者 | Zhang, Y, Daday, C, Gu, R, Cox, C.D, Martinac, B, Groot, B, Walz, T. | | 登録日 | 2020-02-27 | | 公開日 | 2021-02-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Visualization of the mechanosensitive ion channel MscS under membrane tension.
Nature, 590, 2021
|
|
5ADY
 
 | | Cryo-EM structures of the 50S ribosome subunit bound with HflX | | 分子名称: | 23S RRNA, 50S RIBOSOMAL PROTEIN L1, 50S RIBOSOMAL PROTEIN L10, ... | | 著者 | Zhang, Y, Mandava, C.S, Cao, W, Li, X, Zhang, D, Li, N, Zhang, Y, Zhang, X, Qin, Y, Mi, K, Lei, J, Sanyal, S, Gao, N. | | 登録日 | 2015-08-25 | | 公開日 | 2015-10-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Hflx is a Ribosome Splitting Factor Rescuing Stalled Ribosomes Under Stress Conditions
Nat.Struct.Mol.Biol., 22, 2015
|
|
4KMA
 
 | |
4KMH
 
 | |
4KM9
 
 | |
4KMD
 
 | | Crystal structure of Sufud60-Gli1p | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, Sufu, ... | | 著者 | Zhang, Y, Qi, X, Zhang, Z, Wu, G. | | 登録日 | 2013-05-08 | | 公開日 | 2013-11-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural insight into the mutual recognition and regulation between Suppressor of Fused and Gli/Ci.
Nat Commun, 4, 2013
|
|
4KM8
 
 | | Crystal structure of Sufud60 | | 分子名称: | Suppressor of fused homolog | | 著者 | Zhang, Y, Qi, X, Zhang, Z, Wu, G. | | 登録日 | 2013-05-08 | | 公開日 | 2013-11-20 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Structural insight into the mutual recognition and regulation between Suppressor of Fused and Gli/Ci.
Nat Commun, 4, 2013
|
|
1N6G
 
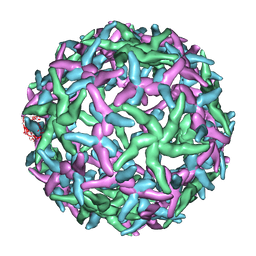 | | The structure of immature Dengue-2 prM particles | | 分子名称: | major envelope protein E | | 著者 | Zhang, Y, Corver, J, Chipman, P.R, Zhang, W, Pletnev, S.V, Sedlak, D, Baker, T.S, Strauss, J.H, Kuhn, R.J, Rossmann, M.G. | | 登録日 | 2002-11-10 | | 公開日 | 2003-06-03 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (16 Å) | | 主引用文献 | Structures of Immature flavivirus particles
EMBO J., 22, 2003
|
|
1NA4
 
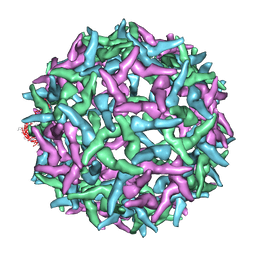 | | The structure of immature Yellow Fever virus particle | | 分子名称: | major envelope protein E | | 著者 | Zhang, Y, Corver, J, Chipman, P.R, Lenches, E, Zhang, W, Pletnev, S.V, Sedlak, D, Baker, T.S, Strauss, J.H, Kuhn, R.J, Rossmann, M.G. | | 登録日 | 2002-11-26 | | 公開日 | 2003-12-09 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY | | 主引用文献 | Structures of immature flavivirus particles
EMBO J., 22, 2003
|
|
1VZX
 
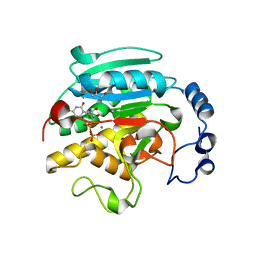 | | Roles of active site tryptophans in substrate binding and catalysis by ALPHA-1,3 GALACTOSYLTRANSFERASE | | 分子名称: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | 著者 | Zhang, Y, Deshpande, A, Xie, Z, Natesh, R, Acharya, K.R, Brew, K. | | 登録日 | 2004-05-28 | | 公開日 | 2004-07-08 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Roles of active site tryptophans in substrate binding and catalysis by alpha-1,3 galactosyltransferase.
Glycobiology, 14, 2004
|
|
