1C5A
 
 | |
2VAH
 
 | | Solution structure of a B-DNA hairpin at low pressure. | | 分子名称: | 5'-D(*AP*GP*GP*AP*TP*CP*CP*TP*UP*TP *TP*GP*GP*AP*TP*CP*CP*T)-3' | | 著者 | Williamson, M.P, Wilton, D.J, Ghosh, M, Chary, K.V.A, Akasaka, K. | | 登録日 | 2007-08-31 | | 公開日 | 2007-09-11 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Change in a B-DNA Helix with Hydrostatic Pressure
Nucleic Acids Res., 36, 2008
|
|
1OA6
 
 | |
1OA5
 
 | |
2BUS
 
 | |
5OAY
 
 | |
1BUS
 
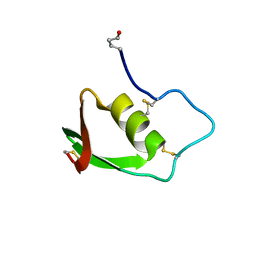 | |
2MCO
 
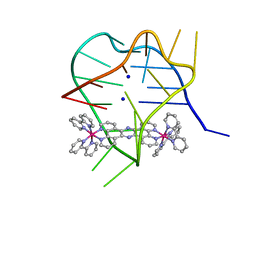 | | Structural studies on dinuclear ruthenium(II) complexes that bind diastereoselectively to an anti-parallel folded human telomere sequence | | 分子名称: | SODIUM ION, human telomere quadruplex, tetrakis(2,2'-bipyridine-kappa~2~N~1~,N~1'~)(mu-tetrapyrido[3,2-a:2',3'-c:3'',2''-h:2''',3'''-j]phenazine-1kappa~2~N~4~,N~5~:2kappa~2~N~13~,N~14~)diruthenium(4+) L enantiomer | | 著者 | Williamson, M.P, Wilson, T, Thomas, J.A, Felix, V, Costa, P.J. | | 登録日 | 2013-08-22 | | 公開日 | 2013-10-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Studies on Dinuclear Ruthenium(II) Complexes That Bind Diastereoselectively to an Antiparallel Folded Human Telomere Sequence.
J.Med.Chem., 56, 2013
|
|
2MCC
 
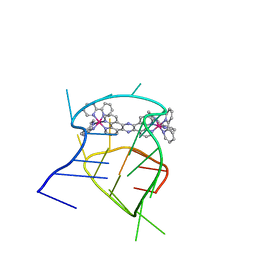 | | Structural studies on dinuclear ruthenium(II) complexes that bind diastereoselectively to an anti-parallel folded human telomere sequence | | 分子名称: | human_telomere_quadruplex, tetrakis(2,2'-bipyridine-kappa~2~N~1~,N~1'~)(mu-tetrapyrido[3,2-a:2',3'-c:3'',2''-h:2''',3'''-j]phenazine-1kappa~2~N~4~,N~5~:2kappa~2~N~13~,N~14~)diruthenium(4+) | | 著者 | Williamson, M.P, Wilson, T, Thomas, J.A, Felix, V, Costa, P.J. | | 登録日 | 2013-08-18 | | 公開日 | 2013-10-02 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Studies on Dinuclear Ruthenium(II) Complexes That Bind Diastereoselectively to an Antiparallel Folded Human Telomere Sequence.
J.Med.Chem., 56, 2013
|
|
2VAI
 
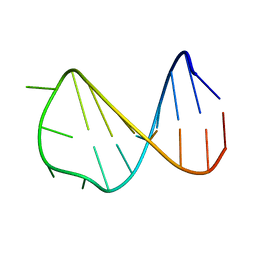 | | Solution structure of a B-DNA hairpin at high pressure | | 分子名称: | 5'-D(*AP*GP*GP*AP*TP*CP*CP*TP*UP*TP *TP*GP*GP*AP*TP*CP*CP*T)-3' | | 著者 | Williamson, M.P, Wilton, D.J, Ghosh, M, Chary, K.V.A, Akasaka, K. | | 登録日 | 2007-08-31 | | 公開日 | 2007-09-11 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural change in a B-DNA helix with hydrostatic pressure.
Nucleic Acids Res., 36, 2008
|
|
2KF6
 
 | |
2KF4
 
 | |
2KF3
 
 | |
2KF5
 
 | |
1KUL
 
 | | GLUCOAMYLASE, GRANULAR STARCH-BINDING DOMAIN, NMR, 5 STRUCTURES | | 分子名称: | GLUCOAMYLASE | | 著者 | Sorimachi, K, Jacks, A.J, Le Gal-Coeffet, M.-F, Williamson, G, Archer, D.B, Williamson, M.P. | | 登録日 | 1996-01-12 | | 公開日 | 1996-07-11 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the granular starch binding domain of glucoamylase from Aspergillus niger by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 259, 1996
|
|
1KUM
 
 | | GLUCOAMYLASE, GRANULAR STARCH-BINDING DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | GLUCOAMYLASE | | 著者 | Sorimachi, K, Jacks, A.J, Le Gal-Coeffet, M.-F, Williamson, G, Archer, D.B, Williamson, M.P. | | 登録日 | 1996-01-12 | | 公開日 | 1996-07-11 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the granular starch binding domain of glucoamylase from Aspergillus niger by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 259, 1996
|
|
6K4I
 
 | | The partially disordered conformation of ubiquitin (Q41N variant) | | 分子名称: | ubiquitin | | 著者 | Wakamoto, T, Ikeya, T, Kitazawa, S, Baxter, N.J, Williamson, M.P, Kitahara, R. | | 登録日 | 2019-05-24 | | 公開日 | 2019-10-30 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Paramagnetic relaxation enhancement-assisted structural characterization of a partially disordered conformation of ubiquitin.
Protein Sci., 28, 2019
|
|
1W8T
 
 | | CBM29-2 mutant K74A complexed with cellulohexaose: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | 分子名称: | NON CATALYTIC PROTEIN 1, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | 登録日 | 2004-09-28 | | 公開日 | 2005-03-22 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
1W7D
 
 | |
1W8W
 
 | | CBM29-2 mutant Y46A: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | 分子名称: | NON-CATALYTIC PROTEIN 1 | | 著者 | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | 登録日 | 2004-09-30 | | 公開日 | 2005-03-22 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
1W8Z
 
 | | CBM29-2 mutant K85A: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | 分子名称: | NON CATALYTIC PROTEIN 1 | | 著者 | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | 登録日 | 2004-10-01 | | 公開日 | 2005-03-22 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
1W90
 
 | | CBM29-2 mutant D114A: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | 分子名称: | 1,2-ETHANEDIOL, NON-CATALYTIC PROTEIN 1, SODIUM ION | | 著者 | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | 登録日 | 2004-10-01 | | 公開日 | 2005-03-18 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
1W9F
 
 | | CBM29-2 mutant R112A: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | 分子名称: | NON CATALYTIC PROTEIN 1 | | 著者 | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | 登録日 | 2004-10-12 | | 公開日 | 2005-03-22 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
1WCU
 
 | | CBM29_1, A Family 29 Carbohydrate Binding Module from Piromyces equi | | 分子名称: | GLYCEROL, NON-CATALYTIC PROTEIN 1 | | 著者 | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davis, G.J, Gilbert, H.J. | | 登録日 | 2004-11-22 | | 公開日 | 2005-03-31 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
2XBD
 
 | | INTERNAL XYLAN BINDING DOMAIN FROM CELLULOMONAS FIMI XYLANASE D, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | XYLANASE D | | 著者 | Simpson, P.J, Bolam, D.N, Cooper, A, Ciruela, A, Hazlewood, G.P, Gilbert, H.J, Williamson, M.P. | | 登録日 | 1998-10-27 | | 公開日 | 1999-07-21 | | 最終更新日 | 2022-03-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A family IIb xylan-binding domain has a similar secondary structure to a homologous family IIa cellulose-binding domain but different ligand specificity.
Structure Fold.Des., 7, 1999
|
|
