7TPJ
 
 | | Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its apo state | | 分子名称: | Fab Heavy (H) Chain, Fab Light (L) Chain, Putative cell surface polysaccharide polymerase/ligase | | 著者 | Ashraf, K.U, Nygaard, R, Vickery, O.N, Erramilli, S.K, Herrera, C.M, McConville, T.H, Petrou, V.I, Giacometti, S.I, Dufrisne, M.B, Nosol, K, Zinkle, A.P, Graham, C.L.B, Loukeris, M, Kloss, B, Skorupinska-Tudek, K, Swiezewska, E, Roper, D, Clarke, O.B, Uhlemann, A.C, Kossiakoff, A.A, Trent, M.S, Stansfeld, P.J, Mancia, F. | | 登録日 | 2022-01-25 | | 公開日 | 2022-04-06 | | 最終更新日 | 2022-04-27 | | 実験手法 | ELECTRON MICROSCOPY (3.46 Å) | | 主引用文献 | Structural basis of lipopolysaccharide maturation by the O-antigen ligase.
Nature, 604, 2022
|
|
7TPG
 
 | | Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its ligand bound state | | 分子名称: | Fab Heavy (H) Chain, Fab Light (L) Chain, GERANYL DIPHOSPHATE, ... | | 著者 | Ashraf, K.U, Nygaard, R, Vickery, O.N, Erramilli, S.K, Herrera, C.M, McConville, T.H, Petrou, V.I, Giacometti, S.I, Dufrisne, M.B, Nosol, K, Zinkle, A.P, Graham, C.L.B, Loukeris, M, Kloss, B, Skorupinska-Tudek, K, Swiezewska, E, Roper, D, Clarke, O.B, Uhlemann, A.C, Kossiakoff, A.A, Trent, M.S, Stansfeld, P.J, Mancia, F. | | 登録日 | 2022-01-25 | | 公開日 | 2022-04-06 | | 最終更新日 | 2022-04-27 | | 実験手法 | ELECTRON MICROSCOPY (3.23 Å) | | 主引用文献 | Structural basis of lipopolysaccharide maturation by the O-antigen ligase.
Nature, 604, 2022
|
|
4OXI
 
 | | Crystal structure of Vibrio cholerae adenylation domain AlmE in complex with glycyl-adenosine-5'-phosphate | | 分子名称: | Enterobactin synthetase component F-related protein, GLYCYL-ADENOSINE-5'-PHOSPHATE | | 著者 | Fage, C.D, Henderson, J.C, Keatinge-Clay, A.T, Trent, M.S. | | 登録日 | 2014-02-05 | | 公開日 | 2014-12-31 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.261 Å) | | 主引用文献 | Antimicrobial peptide resistance of Vibrio cholerae results from an LPS modification pathway related to nonribosomal peptide synthetases.
Acs Chem.Biol., 9, 2014
|
|
4TN0
 
 | | Crystal Structure of the C-terminal Periplasmic Domain of Phosphoethanolamine Transferase EptC from Campylobacter jejuni | | 分子名称: | UPF0141 protein yjdB, ZINC ION | | 著者 | Fage, C.D, Brown, D, Boll, J.M, Keatinge-Clay, A.T, Trent, M.S. | | 登録日 | 2014-06-02 | | 公開日 | 2014-10-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystallographic study of the phosphoethanolamine transferase EptC required for polymyxin resistance and motility in Campylobacter jejuni.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3FID
 
 | | LpxR from Salmonella typhimurium | | 分子名称: | GLYCEROL, PENTAETHYLENE GLYCOL MONODECYL ETHER, Putative outer membrane protein (LpxR), ... | | 著者 | Rutten, L, Gros, P. | | 登録日 | 2008-12-11 | | 公開日 | 2009-02-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Active-site architecture and catalytic mechanism of the lipid A deacylase LpxR of Salmonella typhimurium
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5F15
 
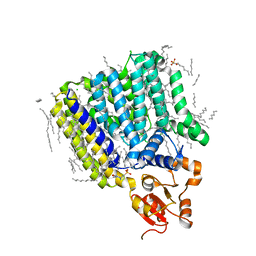 | | Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-amino-4-deoxy-L-arabinose (L-Ara4N) transferase, CHLORIDE ION, ... | | 著者 | Petrou, V.I, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | 登録日 | 2015-11-30 | | 公開日 | 2016-02-17 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation.
Science, 351, 2016
|
|
5EZM
 
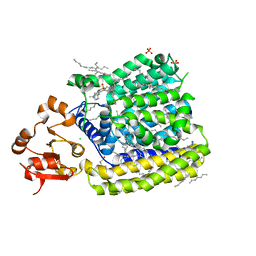 | | Crystal Structure of ArnT from Cupriavidus metallidurans in the apo state | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferases of PMT family, CHLORIDE ION, ... | | 著者 | Petrou, V.I, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | 登録日 | 2015-11-26 | | 公開日 | 2016-02-17 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation.
Science, 351, 2016
|
|
