3A62
 
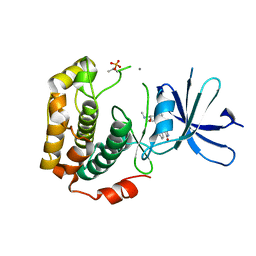 | | Crystal structure of phosphorylated p70S6K1 | | 分子名称: | MANGANESE (II) ION, Ribosomal protein S6 kinase beta-1, STAUROSPORINE | | 著者 | Sunami, T, Byrne, N, Diehl, R.E, Funabashi, K, Hall, D.L, Ikuta, M, Patel, S.B, Shipman, J.M, Smith, R.F, Takahashi, I, Zugay-Murphy, J, Iwasawa, Y, Lumb, K.J, Munshi, S.K, Sharma, S. | | 登録日 | 2009-08-18 | | 公開日 | 2009-10-27 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural basis of human p70 ribosomal S6 kinase-1 regulation by activation loop phosphorylation.
J.Biol.Chem., 285, 2010
|
|
3A61
 
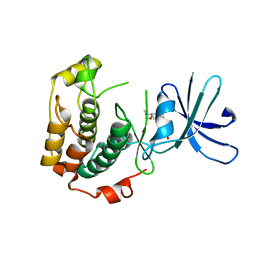 | | Crystal structure of unphosphorylated p70S6K1 (Form II) | | 分子名称: | Ribosomal protein S6 kinase beta-1, STAUROSPORINE | | 著者 | Sunami, T, Byrne, N, Diehl, R.E, Funabashi, K, Hall, D.L, Ikuta, M, Patel, S.B, Shipman, J.M, Smith, R.F, Takahashi, I, Zugay-Murphy, J, Iwasawa, Y, Lumb, K.J, Munshi, S.K, Sharma, S. | | 登録日 | 2009-08-18 | | 公開日 | 2009-10-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.43 Å) | | 主引用文献 | Structural basis of human p70 ribosomal S6 kinase-1 regulation by activation loop phosphorylation.
J.Biol.Chem., 285, 2010
|
|
3A60
 
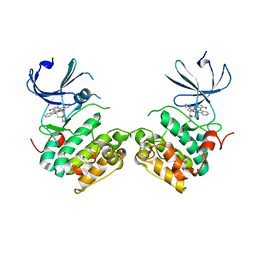 | | Crystal structure of unphosphorylated p70S6K1 (Form I) | | 分子名称: | Ribosomal protein S6 kinase beta-1, STAUROSPORINE | | 著者 | Sunami, T, Byrne, N, Diehl, R.E, Funabashi, K, Hall, D.L, Ikuta, M, Patel, S.B, Shipman, J.M, Smith, R.F, Takahashi, I, Zugay-Murphy, J, Iwasawa, Y, Lumb, K.J, Munshi, S.K, Sharma, S. | | 登録日 | 2009-08-17 | | 公開日 | 2009-10-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis of human p70 ribosomal S6 kinase-1 regulation by activation loop phosphorylation.
J.Biol.Chem., 285, 2010
|
|
3NBS
 
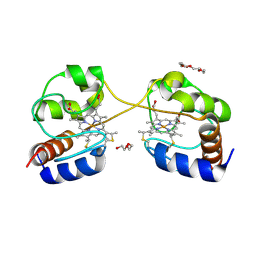 | | Crystal structure of dimeric cytochrome c from horse heart | | 分子名称: | Cytochrome c, DI(HYDROXYETHYL)ETHER, HEME C, ... | | 著者 | Taketa, M, Komori, H, Hirota, S, Higuchi, Y. | | 登録日 | 2010-06-04 | | 公開日 | 2010-07-14 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Cytochrome c polymerization by successive domain swapping at the C-terminal helix
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3NBT
 
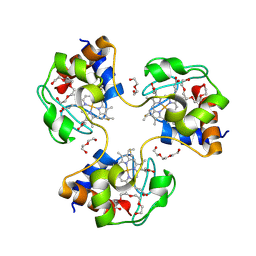 | | Crystal structure of trimeric cytochrome c from horse heart | | 分子名称: | Cytochrome c, DI(HYDROXYETHYL)ETHER, HEME C, ... | | 著者 | Taketa, M, Komori, H, Hirota, S, Higuchi, Y. | | 登録日 | 2010-06-04 | | 公開日 | 2010-07-14 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Cytochrome c polymerization by successive domain swapping at the C-terminal helix
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1GIJ
 
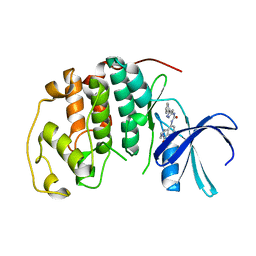 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE CDK4 INHIBITOR | | 分子名称: | 1-(5-OXO-2,3,5,9B-TETRAHYDRO-1H-PYRROLO[2,1-A]ISOINDOL-9-YL)-3-(5-PYRROLIDIN-2-YL-1H-PYRAZOL-3-YL)-UREA, CELL DIVISION PROTEIN KINASE 2 | | 著者 | Ikuta, M, Nishimura, S. | | 登録日 | 2001-02-06 | | 公開日 | 2002-02-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystallographic approach to identification of cyclin-dependent kinase 4 (CDK4)-specific inhibitors by using CDK4 mimic CDK2 protein.
J.Biol.Chem., 276, 2002
|
|
1GII
 
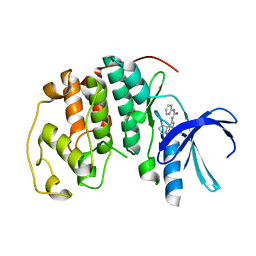 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE CDK4 INHIBITOR | | 分子名称: | 1-(5-OXO-2,3,5,9B-TETRAHYDRO-1H-PYRROLO[2,1-A]ISOINDOL-9-YL)-3-PYRIDIN-2-YL-UREA, CELL DIVISION PROTEIN KINASE 2 | | 著者 | Ikuta, M, Nishimura, S. | | 登録日 | 2001-02-06 | | 公開日 | 2002-02-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystallographic approach to identification of cyclin-dependent kinase 4 (CDK4)-specific inhibitors by using CDK4 mimic CDK2 protein.
J.Biol.Chem., 276, 2001
|
|
1GIH
 
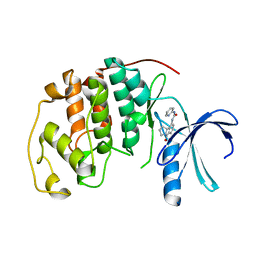 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE CDK4 INHIBITOR | | 分子名称: | 1-(5-OXO-2,3,5,9B-TETRAHYDRO-1H-PYRROLO[2,1-A]ISOINDOL-9-YL)-3-PYRIDIN-2-YL-UREA, CELL DIVISION PROTEIN KINASE 2 | | 著者 | Ikuta, M, Nishimura, S. | | 登録日 | 2001-02-06 | | 公開日 | 2002-02-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystallographic approach to identification of cyclin-dependent kinase 4 (CDK4)-specific inhibitors by using CDK4 mimic CDK2 protein.
J.Biol.Chem., 276, 2001
|
|
7VKA
 
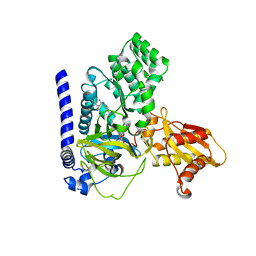 | | Crystal Structure of GH3.6 in complex with an inhibitor | | 分子名称: | ADENOSINE MONOPHOSPHATE, GLYCEROL, Indole-3-acetic acid-amido synthetase GH3.6, ... | | 著者 | Wang, N, Luo, M, Bao, H, Huang, H. | | 登録日 | 2021-09-29 | | 公開日 | 2022-08-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.401 Å) | | 主引用文献 | Chemical genetic screening identifies nalacin as an inhibitor of GH3 amido synthetase for auxin conjugation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5ZHT
 
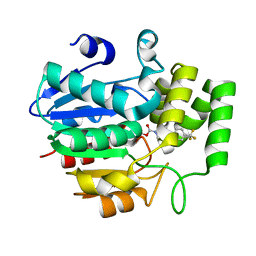 | | Crystal structure of OsD14 in complex with covalently bound KK073 | | 分子名称: | (1H-1,2,3-triazol-1-yl){4-[4-(trifluoromethyl)phenyl]piperazin-1-yl}methanone, Strigolactone esterase D14 | | 著者 | Hirabayashi, K, Miyakawa, T, Tanokura, M. | | 登録日 | 2018-03-13 | | 公開日 | 2018-11-21 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.532 Å) | | 主引用文献 | Triazole Ureas Covalently Bind to Strigolactone Receptor and Antagonize Strigolactone Responses.
Mol Plant, 12, 2019
|
|
5ZHR
 
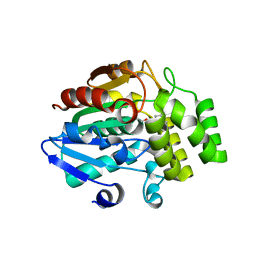 | | Crystal structure of OsD14 in complex with covalently bound KK094 | | 分子名称: | (2,3-dihydro-1H-indol-1-yl)(1H-1,2,3-triazol-1-yl)methanone, Strigolactone esterase D14 | | 著者 | Hirabayashi, K, Miyakawa, T, Tanokura, M. | | 登録日 | 2018-03-13 | | 公開日 | 2018-11-21 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Triazole Ureas Covalently Bind to Strigolactone Receptor and Antagonize Strigolactone Responses.
Mol Plant, 12, 2019
|
|
5ZHS
 
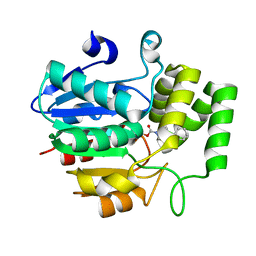 | | Crystal structure of OsD14 in complex with covalently bound KK052 | | 分子名称: | (4-phenylpiperazin-1-yl)(1H-1,2,3-triazol-1-yl)methanone, Strigolactone esterase D14 | | 著者 | Hirabayashi, K, Miyakawa, T, Tanokura, M. | | 登録日 | 2018-03-13 | | 公開日 | 2018-11-21 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Triazole Ureas Covalently Bind to Strigolactone Receptor and Antagonize Strigolactone Responses.
Mol Plant, 12, 2019
|
|
3A8W
 
 | | Crystal Structure of PKCiota kinase domain | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Protein kinase C iota type, SULFATE ION | | 著者 | Takimura, T, Kamata, K. | | 登録日 | 2009-10-11 | | 公開日 | 2010-05-05 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structures of the PKC-iota kinase domain in its ATP-bound and apo forms reveal defined structures of residues 533-551 in the C-terminal tail and their roles in ATP binding
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3A8X
 
 | | Crystal Structure of PKCiota kinase domain | | 分子名称: | Protein kinase C iota type, SULFATE ION | | 著者 | Takimura, T, Kamata, K. | | 登録日 | 2009-10-11 | | 公開日 | 2010-05-05 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structures of the PKC-iota kinase domain in its ATP-bound and apo forms reveal defined structures of residues 533-551 in the C-terminal tail and their roles in ATP binding
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3VXW
 
 | |
