6JQN
 
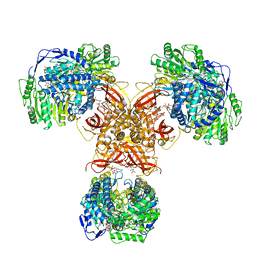 | | Structure of PaaZ, a bifunctional enzyme in complex with NADP+ and OCoA | | 分子名称: | Bifunctional protein PaaZ, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OCTANOYL-COENZYME A | | 著者 | Gakher, L, Vinothkumar, K.R, Katagihallimath, N, Sowdhamini, R, Sathyanarayanan, N, Cannone, G. | | 登録日 | 2019-03-31 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun, 10, 2019
|
|
6JQO
 
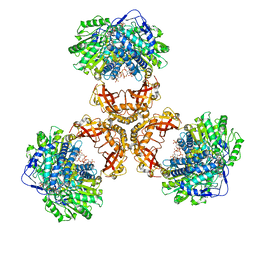 | | Structure of PaaZ, a bifunctional enzyme in complex with NADP+ and CCoA | | 分子名称: | Bifunctional protein PaaZ, CROTONYL COENZYME A, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Gakher, L, Vinothkumar, K.R, Katagihallimath, N, Sowdhamini, R, Sathyanarayanan, N, Cannone, G. | | 登録日 | 2019-03-31 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun, 10, 2019
|
|
6JQL
 
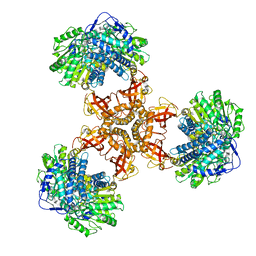 | | Structure of PaaZ, a bifunctional enzyme | | 分子名称: | Bifunctional protein PaaZ | | 著者 | Gakher, L, Vinothkumar, K.R, Katagihallimath, N, Sowdhamini, R, Sathyanarayanan, N, Cannone, G. | | 登録日 | 2019-03-31 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun, 10, 2019
|
|
6JQM
 
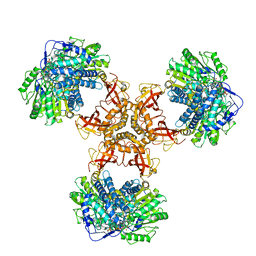 | | Structure of PaaZ with NADPH | | 分子名称: | Bifunctional protein PaaZ, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Gakher, L, Vinothkumar, K.R, Katagihallimath, N, Sowdhamini, R, Sathyanarayanan, N, Cannone, G. | | 登録日 | 2019-03-31 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun, 10, 2019
|
|
6JKU
 
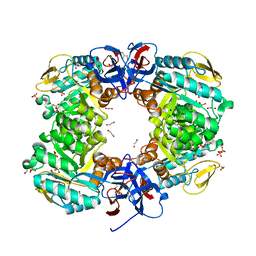 | | Crystal structure of N-acetylglucosamine-6-phosphate deacetylase from Pasteurella Multocida | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, N-acetylglucosamine-6-phosphate deacetylase, ... | | 著者 | Manjunath, L, Bose, S, Subramanian, R. | | 登録日 | 2019-03-01 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Quaternary variations in the structural assembly of N-acetylglucosamine-6-phosphate deacetylase from Pasteurella multocida.
Proteins, 2020
|
|
8C6G
 
 | |
8C68
 
 | |
8C6E
 
 | |
