2MOV
 
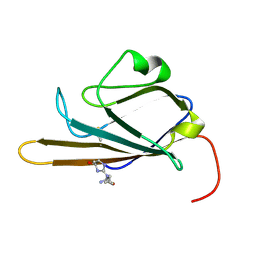 | | Receptor for Advanced Glycation End Products (RAGE) Specifically Recognizes Methylglyoxal Derived AGEs. | | 分子名称: | Advanced glycosylation end product-specific receptor, N~5~-[(5R)-5-methyl-4-oxo-4,5-dihydro-1H-imidazol-2-yl]-L-ornithine | | 著者 | Shekhtman, A, Xue, J, Ray, R, Singer, D, Bohme, D, Burz, D.S, Rai, V, Hoffman, R. | | 登録日 | 2014-05-05 | | 公開日 | 2014-06-25 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Receptor for Advanced Glycation End Products (RAGE) Specifically Recognizes Methylglyoxal-Derived AGEs.
Biochemistry, 53, 2014
|
|
2L7U
 
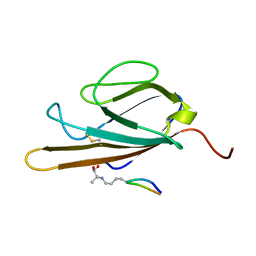 | | Structure of CEL-PEP-RAGE V domain complex | | 分子名称: | Advanced glycosylation end product-specific receptor, Serum albumin peptide | | 著者 | Xue, J, Rai, V, Schmidt, A, Frolov, S, Reverdatto, S, Singer, D, Chabierski, S, Xie, J, Burz, D, Shekhtman, A, Hoffman, R. | | 登録日 | 2010-12-23 | | 公開日 | 2011-05-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Advanced glycation end product recognition by the receptor for AGEs.
Structure, 19, 2011
|
|
4K7M
 
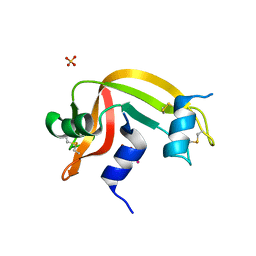 | |
4K7L
 
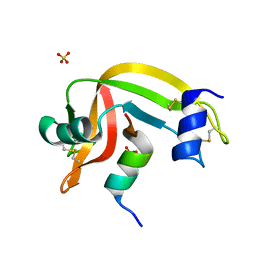 | | Crystal structure of RNase S variant (K7C/Q11C) | | 分子名称: | Ribonuclease pancreatic, SULFATE ION | | 著者 | Genz, M, Straeter, N. | | 登録日 | 2013-04-17 | | 公開日 | 2014-05-14 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Crystal structure of RNase S with a [Hg(Cys2)] metal center in the S-peptide as a template for structure-based design of artificial metalloenzymes using peptide-protein complementation
To be Published
|
|
4O36
 
 | | Semisynthetic RNase S1-15-H7/11-Q10 | | 分子名称: | CHLORIDE ION, Ribonuclease pancreatic, S-peptide, ... | | 著者 | Genz, M, Strater, N. | | 登録日 | 2013-12-18 | | 公開日 | 2014-05-07 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | An Artificial Imine Reductase based on the Ribonuclease S scaffold
Chem.Cat.Chem, 2014
|
|
4O37
 
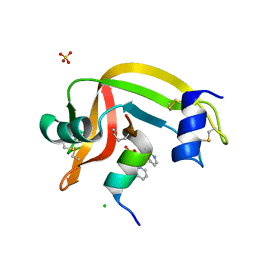 | | seminsynthetic RNase S1-15-3Pl-7/11 | | 分子名称: | CHLORIDE ION, Ribonuclease pancreatic, S-peptide, ... | | 著者 | Genz, M, Strater, N. | | 登録日 | 2013-12-18 | | 公開日 | 2014-05-07 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | An Artificial Imine Reductase based on the Ribonuclease S Scaffold
Chem.Cat.Chem, 2014
|
|
3O5B
 
 | | Crystal structure of dimeric KlHxk1 in crystal form VII with glucose bound (open state) | | 分子名称: | Hexokinase, SULFATE ION, beta-D-glucopyranose | | 著者 | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | 登録日 | 2010-07-28 | | 公開日 | 2010-10-13 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3O08
 
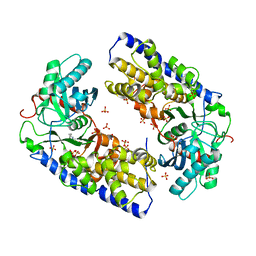 | | Crystal structure of dimeric KlHxk1 in crystal form I | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hexokinase, SULFATE ION | | 著者 | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | 登録日 | 2010-07-19 | | 公開日 | 2010-10-13 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3O4W
 
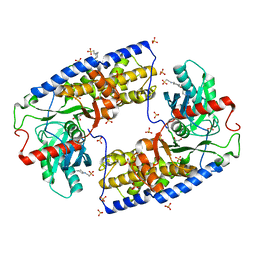 | | Crystal structure of dimeric KlHxk1 in crystal form IV | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Hexokinase, ... | | 著者 | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | 登録日 | 2010-07-27 | | 公開日 | 2010-10-13 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3O8M
 
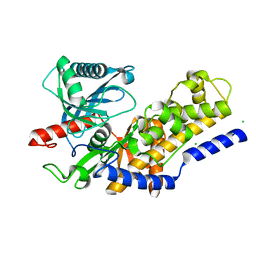 | | Crystal structure of monomeric KlHxk1 in crystal form XI with glucose bound (closed state) | | 分子名称: | CHLORIDE ION, Hexokinase, alpha-D-glucopyranose, ... | | 著者 | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | 登録日 | 2010-08-03 | | 公開日 | 2010-10-13 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3O1B
 
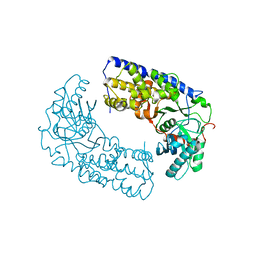 | | CRYSTAL STRUCTURE OF DIMERIC KLHXK1 IN CRYSTAL FORM II | | 分子名称: | Hexokinase | | 著者 | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | 登録日 | 2010-07-21 | | 公開日 | 2010-10-13 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3O1W
 
 | | Crystal structure of dimeric KlHxk1 in crystal form III | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Hexokinase, ... | | 著者 | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | 登録日 | 2010-07-22 | | 公開日 | 2010-10-13 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3O6W
 
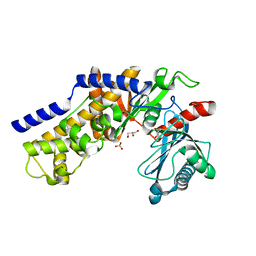 | | Crystal structure of monomeric KlHxk1 in crystal form VIII (open state) | | 分子名称: | GLYCEROL, Hexokinase, PHOSPHATE ION | | 著者 | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | 登録日 | 2010-07-29 | | 公開日 | 2010-10-13 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3O80
 
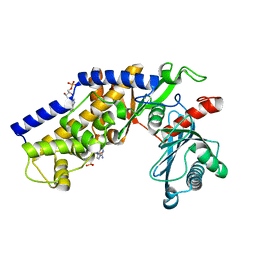 | | Crystal structure of monomeric KlHxk1 in crystal form IX (open state) | | 分子名称: | Hexokinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | 登録日 | 2010-08-02 | | 公開日 | 2010-10-13 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
2NBQ
 
 | |
