5EI0
 
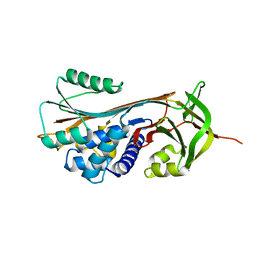 | | Structure of RCL-cleaved vaspin (serpinA12) | | 分子名称: | Serpin A12 | | 著者 | Pippel, J, Kuettner, B.E, Ulbricht, D, Daberger, J, Schultz, S, Heiker, J.T, Strater, N. | | 登録日 | 2015-10-29 | | 公開日 | 2015-11-11 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of cleaved vaspin (serpinA12).
Biol.Chem., 397, 2016
|
|
1BS5
 
 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM | | 分子名称: | PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ZINC ION | | 著者 | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | 登録日 | 1998-09-01 | | 公開日 | 1999-08-27 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BS8
 
 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM IN COMPLEX WITH TRIPEPTIDE MET-ALA-SER | | 分子名称: | PROTEIN (MET-ALA-SER), PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ... | | 著者 | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | 登録日 | 1998-09-01 | | 公開日 | 1999-08-27 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BS7
 
 | | PEPTIDE DEFORMYLASE AS NI2+ CONTAINING FORM | | 分子名称: | NICKEL (II) ION, PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION | | 著者 | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | 登録日 | 1998-09-01 | | 公開日 | 1999-08-27 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure of peptide deformylase and identification of the substrate binding site.
J.Biol.Chem., 273, 1998
|
|
1BS4
 
 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM (NATIVE) IN COMPLEX WITH INHIBITOR POLYETHYLENE GLYCOL | | 分子名称: | NONAETHYLENE GLYCOL, PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ... | | 著者 | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | 登録日 | 1998-09-01 | | 公開日 | 1999-08-27 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BS6
 
 | | PEPTIDE DEFORMYLASE AS NI2+ CONTAINING FORM IN COMPLEX WITH TRIPEPTIDE MET-ALA-SER | | 分子名称: | NICKEL (II) ION, PROTEIN (MET-ALA-SER), PROTEIN (PEPTIDE DEFORMYLASE), ... | | 著者 | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | 登録日 | 1998-09-01 | | 公開日 | 1999-08-27 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BSZ
 
 | | PEPTIDE DEFORMYLASE AS FE2+ CONTAINING FORM (NATIVE) IN COMPLEX WITH INHIBITOR POLYETHYLENE GLYCOL | | 分子名称: | FE (III) ION, NONAETHYLENE GLYCOL, PROTEIN (PEPTIDE DEFORMYLASE), ... | | 著者 | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | 登録日 | 1998-09-01 | | 公開日 | 1999-08-26 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
4IF8
 
 | | Structure Of Vaspin | | 分子名称: | SULFATE ION, Serpin A12 | | 著者 | Kuettner, E.B, Strater, N, Heiker, J.T, Kloting, N, Kovacs, P, Schultz, S, Kern, M, Stumvoll, M, Bluher, M, Beck-Sickinger, A.G. | | 登録日 | 2012-12-14 | | 公開日 | 2013-02-13 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Vaspin inhibits kallikrein 7 by serpin mechanism
Cell.Mol.Life Sci., 70, 2013
|
|
3PFL
 
 | | CRYSTAL STRUCTURE OF PFL FROM E.COLI IN COMPLEX WITH SUBSTRATE ANALOGUE OXAMATE | | 分子名称: | OXAMIC ACID, PROTEIN (FORMATE ACETYLTRANSFERASE 1) | | 著者 | Becker, A, Fritz-Wolf, K, Kabsch, W, Knappe, J, Schultz, S, Wagner, A.F.V. | | 登録日 | 1999-05-14 | | 公開日 | 2000-05-31 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase.
Nat.Struct.Biol., 6, 1999
|
|
2PFL
 
 | | CRYSTAL STRUCTURE OF PFL FROM E.COLI | | 分子名称: | CHLORIDE ION, PROTEIN (PYRUVATE FORMATE-LYASE), SODIUM ION | | 著者 | Becker, A, Fritz-Wolf, K, Kabsch, W, Knappe, J, Schultz, S, Wagner, A.F.V. | | 登録日 | 1999-05-26 | | 公開日 | 1999-12-15 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase.
Nat.Struct.Biol., 6, 1999
|
|
1ICJ
 
 | | PDF PROTEIN IS CRYSTALLIZED AS NI2+ CONTAINING FORM, COCRYSTALLIZED WITH INHIBITOR POLYETHYLENE GLYCOL (PEG) | | 分子名称: | NICKEL (II) ION, NONAETHYLENE GLYCOL, PEPTIDE DEFORMYLASE, ... | | 著者 | Becker, A, Schlichting, I, Kabsch, W, Schultz, S, Wagner, A.F.V. | | 登録日 | 1998-03-12 | | 公開日 | 1999-03-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of peptide deformylase and identification of the substrate binding site.
J.Biol.Chem., 273, 1998
|
|
4Y3K
 
 | | Structure of Vaspin mutant E379S | | 分子名称: | 1,2-ETHANEDIOL, SULFATE ION, Serpin A12 | | 著者 | Pippel, J, Strater, N, Ulbricht, D, Schultz, S, Meier, R, Heiker, J.T. | | 登録日 | 2015-02-10 | | 公開日 | 2015-08-12 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | A unique serpin P1' glutamate and a conserved beta-sheet C arginine are key residues for activity, protease recognition and stability of serpinA12 (vaspin).
Biochem.J., 470, 2015
|
|
4Y40
 
 | | Structure of Vaspin mutant D305C V383C | | 分子名称: | 1,2-ETHANEDIOL, SULFATE ION, Serpin A12 | | 著者 | Pippel, J, Strater, N, Ulbricht, D, Schultz, S, Meier, R, Heiker, J.T. | | 登録日 | 2015-02-10 | | 公開日 | 2015-08-12 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | A unique serpin P1' glutamate and a conserved beta-sheet C arginine are key residues for activity, protease recognition and stability of serpinA12 (vaspin).
Biochem.J., 470, 2015
|
|
1CM5
 
 | | CRYSTAL STRUCTURE OF C418A,C419A MUTANT OF PFL FROM E.COLI | | 分子名称: | CARBONATE ION, PROTEIN (PYRUVATE FORMATE-LYASE), SODIUM ION | | 著者 | Becker, A, Fritz-Wolf, K, Kabsch, W, Knappe, J, Schultz, S, Wagner, A.F.V. | | 登録日 | 1999-05-14 | | 公開日 | 1999-12-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase.
Nat.Struct.Biol., 6, 1999
|
|
5HG9
 
 | | EGFR (L858R, T790M, V948R) in complex with 1-[(3R,4R)-3-[({2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-(trifluoromethyl)pyrrolidin-1-yl]prop-2-en-1-one | | 分子名称: | 1-[(3R,4R)-3-[({2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-(trifluoromethyl)pyrrolidin-1-yl]propan-1-one, Epidermal growth factor receptor, GLYCEROL, ... | | 著者 | Gajiwala, K.S. | | 登録日 | 2016-01-08 | | 公開日 | 2016-02-03 | | 最終更新日 | 2016-03-23 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Discovery of 1-{(3R,4R)-3-[({5-Chloro-2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-methoxypyrrolidin-1-yl}prop-2-en-1-one (PF-06459988), a Potent, WT Sparing, Irreversible Inhibitor of T790M-Containing EGFR Mutants.
J.Med.Chem., 59, 2016
|
|
5HG8
 
 | | EGFR (L858R, T790M, V948R) in complex with N-[3-({2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)phenyl]prop-2-enamide | | 分子名称: | Epidermal growth factor receptor, GLYCEROL, N-[3-({2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)phenyl]propanamide, ... | | 著者 | Gajiwala, K.S. | | 登録日 | 2016-01-08 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Discovery of 1-{(3R,4R)-3-[({5-Chloro-2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-methoxypyrrolidin-1-yl}prop-2-en-1-one (PF-06459988), a Potent, WT Sparing, Irreversible Inhibitor of T790M-Containing EGFR Mutants.
J.Med.Chem., 59, 2016
|
|
5HG5
 
 | | EGFR (L858R, T790M, V948R) in complex with N-{3-[(2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-7H-pyrrolo[2,3-d]pyrimidin-4-yl)oxy]phenyl}prop-2-enamide | | 分子名称: | Epidermal growth factor receptor, GLYCEROL, N-{3-[(2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-7H-pyrrolo[2,3-d]pyrimidin-4-yl)oxy]phenyl}propanamide, ... | | 著者 | Gajiwala, K.S. | | 登録日 | 2016-01-08 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Discovery of 1-{(3R,4R)-3-[({5-Chloro-2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-methoxypyrrolidin-1-yl}prop-2-en-1-one (PF-06459988), a Potent, WT Sparing, Irreversible Inhibitor of T790M-Containing EGFR Mutants.
J.Med.Chem., 59, 2016
|
|
5HG7
 
 | | EGFR (L858R, T790M, V948R) in complex with 1-{(3R,4R)-3-[5-Chloro-2-(1-methyl-1H-pyrazol-4-ylamino)-7H-pyrrolo[2,3-d]pyrimidin-4-yloxymethyl]-4-methoxy-pyrrolidin-1-yl}propenone (PF-06459988) | | 分子名称: | 1-{(3R,4R)-3-[({5-chloro-2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-methoxypyrrolidin-1-yl}propan-1-one, Epidermal growth factor receptor, SULFATE ION | | 著者 | Gajiwala, K.S. | | 登録日 | 2016-01-08 | | 公開日 | 2016-01-27 | | 最終更新日 | 2016-03-23 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Discovery of 1-{(3R,4R)-3-[({5-Chloro-2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-methoxypyrrolidin-1-yl}prop-2-en-1-one (PF-06459988), a Potent, WT Sparing, Irreversible Inhibitor of T790M-Containing EGFR Mutants.
J.Med.Chem., 59, 2016
|
|
8FP3
 
 | | PKCeta kinase domain in complex with compound 11 | | 分子名称: | (3P)-3-{4-[(3R,5S)-3-amino-5-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile, Protein kinase C eta type | | 著者 | Johnson, E. | | 登録日 | 2023-01-03 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8FH4
 
 | | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with 3-[6-chloro-4-(9-methyl-1-oxa-4,9-diazaspiro[5.5]undec-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile | | 分子名称: | (3P)-3-[6-chloro-4-(9-methyl-1-oxa-4,9-diazaspiro[5.5]undecan-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile, Mitogen-activated protein kinase kinase kinase kinase 1, SULFATE ION | | 著者 | McTigue, M, Johnson, E, Cronin, C. | | 登録日 | 2022-12-13 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.827 Å) | | 主引用文献 | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8FKO
 
 | | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with 3-{4-[(2S,5R)-5-Amino-2-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile | | 分子名称: | (3P)-3-{4-[(2S,5R)-5-amino-2-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile, Mitogen-activated protein kinase kinase kinase kinase 1 | | 著者 | McTigue, M, Johnson, E, Cronin, C. | | 登録日 | 2022-12-21 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.104 Å) | | 主引用文献 | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8FJZ
 
 | | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with 3-{4-[(3R,5S)-3-Amino-5-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile | | 分子名称: | (3P)-3-{4-[(3R,5S)-3-amino-5-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile, Mitogen-activated protein kinase kinase kinase kinase 1 | | 著者 | McTigue, M, Johnson, E, Cronin, C. | | 登録日 | 2022-12-20 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.897 Å) | | 主引用文献 | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8FP1
 
 | | PKCeta kinase domain in complex with compound 2 | | 分子名称: | (3P)-3-[6-chloro-4-(9-methyl-1-oxa-4,9-diazaspiro[5.5]undecan-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile, Protein kinase C eta type | | 著者 | Johnson, E. | | 登録日 | 2023-01-03 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-04-26 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
3HEK
 
 | |
5UG9
 
 | | Crystal structure of the EGFR kinase domain (L858R, T790M, V948R) in complex with a covalent inhibitor N-[(3R,4R)-4-fluoro-1-{6-[(3-methoxy-1-methyl-1H-pyrazol-4-yl)amino]-9-(propan-2-yl)-9H-purin-2-yl}pyrrolidin-3-yl]propanamide | | 分子名称: | 1,2-ETHANEDIOL, Epidermal growth factor receptor, GLYCEROL, ... | | 著者 | Gajiwala, K.S, Ferre, R.A. | | 登録日 | 2017-01-07 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | Discovery of N-((3R,4R)-4-Fluoro-1-(6-((3-methoxy-1-methyl-1H-pyrazol-4-yl)amino)-9-methyl-9H-purin-2-yl)pyrrolidine-3-yl)acrylamide (PF-06747775) through Structure-Based Drug Design: A High Affinity Irreversible Inhibitor Targeting Oncogenic EGFR Mutants with Selectivity over Wild-Type EGFR.
J. Med. Chem., 60, 2017
|
|
