4W8L
 
 | |
4XUR
 
 | |
4XUQ
 
 | |
4XUO
 
 | |
4XUT
 
 | | Structure of the CBM22-2 xylan-binding domain in complex with 1,3:1,4 Beta-glucotetraose B from Paenibacillus barcinonensis Xyn10C | | 分子名称: | CALCIUM ION, Endo-1,4-beta-xylanase C, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Sainz-Polo, M.A, Sanz-Aparicio, J. | | 登録日 | 2015-01-26 | | 公開日 | 2015-06-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Exploring Multimodularity in Plant Cell Wall Deconstruction: STRUCTURAL AND FUNCTIONAL ANALYSIS OF Xyn10C CONTAINING THE CBM22-1-CBM22-2 TANDEM.
J.Biol.Chem., 290, 2015
|
|
4XUP
 
 | |
4XUN
 
 | |
4EQV
 
 | |
3U14
 
 | | Structure of D50A-fructofuranosidase from Schwanniomyces occidentalis complexed with inulin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fructofuranosidase, beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose | | 著者 | Sainz-Polo, M.A, Sanz-Aparicio, J. | | 登録日 | 2011-09-29 | | 公開日 | 2012-04-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Structural and kinetic insights reveal that the amino acid pair GLN228/ASN254 modulates the transfructosylating specificity of Schwanniomyces occidentalis beta-fructofuranosidase, an enzyme that produces prebiotics.
J.Biol.Chem., 287, 2012
|
|
3U75
 
 | |
4QB6
 
 | |
4QB1
 
 | |
4QAW
 
 | |
4QB2
 
 | |
6H8L
 
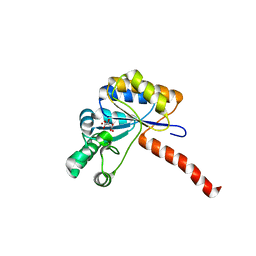 | | Structure of peptidoglycan deacetylase PdaC from Bacillus subtilis | | 分子名称: | L(+)-TARTARIC ACID, Peptidoglycan-N-acetylmuramic acid deacetylase PdaC, ZINC ION | | 著者 | Sainz-Polo, M.A, Grifoll-Romero, L, Albesa-Jove, D, Planas, A, Guerin, M.E. | | 登録日 | 2018-08-02 | | 公開日 | 2019-11-13 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Structure-function relationships underlying the dualN-acetylmuramic andN-acetylglucosamine specificities of the bacterial peptidoglycan deacetylase PdaC.
J.Biol.Chem., 294, 2019
|
|
6H8N
 
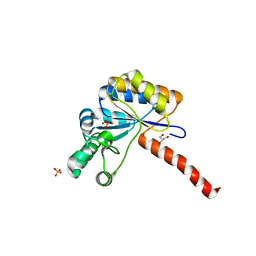 | | Structure of peptidoglycan deacetylase PdaC from Bacillus subtilis - mutant D285S | | 分子名称: | GLYCEROL, PHOSPHATE ION, Peptidoglycan-N-acetylmuramic acid deacetylase PdaC, ... | | 著者 | Sainz-Polo, M.A, Grifoll-Romero, L, Albesa-Jove, D, Planas, A, Guerin, M.E. | | 登録日 | 2018-08-02 | | 公開日 | 2019-11-13 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.26 Å) | | 主引用文献 | Structure-function relationships underlying the dualN-acetylmuramic andN-acetylglucosamine specificities of the bacterial peptidoglycan deacetylase PdaC.
J.Biol.Chem., 294, 2019
|
|
6YHV
 
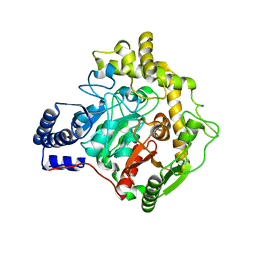 | | Structural insights into Pseudomonas aeruginosa Type six secretion system exported effector 8: unliganded Tse8 | | 分子名称: | COPPER (II) ION, Tse8 | | 著者 | Sainz-Polo, M.A, Capuni, R, Pretre, G, Gonzalez-Magana, A, Lucas, M, Altuna, J, Montanchez, I, Fucini, P, Albesa-Jove, D. | | 登録日 | 2020-03-31 | | 公開日 | 2020-11-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.893 Å) | | 主引用文献 | Structural insights into Pseudomonas aeruginosaType six secretion system exported effector 8.
J.Struct.Biol., 212, 2020
|
|
6TE4
 
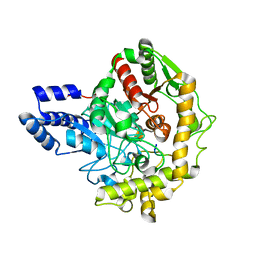 | | Structural insights into Pseudomonas aeruginosa Type six secretion system exported effector 8: Tse8 in complex with a peptide | | 分子名称: | Pro-Pro-Leu-Ala-Ser-Lys, Tse8 | | 著者 | Sainz-Polo, M.A, Capuni, R, Lucas, M, Altuna, J, Fucini, P, Montanchez, I, Albesa-Jove, D. | | 登録日 | 2019-11-11 | | 公開日 | 2020-11-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Structural insights into Pseudomonas aeruginosaType six secretion system exported effector 8.
J.Struct.Biol., 212, 2020
|
|
5NRD
 
 | | A Native Ternary Complex of Alpha-1,3-Galactosyltransferase (a-3GalT) Supports a Conserved Reaction Mechanism for Retaining Glycosyltransferases - alpha-3GalT in complex with Co2+, UDP-Gal and lactose - a3GalT-Co2+-UDP-Gal-LAT-2 | | 分子名称: | COBALT (II) ION, GALACTOSE-URIDINE-5'-DIPHOSPHATE, GLYCEROL, ... | | 著者 | Albesa-Jove, D, Marina, A, Sainz-Polo, M.A, Guerin, M.E. | | 登録日 | 2017-04-22 | | 公開日 | 2017-10-11 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Structural Snapshots of alpha-1,3-Galactosyltransferase with Native Substrates: Insight into the Catalytic Mechanism of Retaining Glycosyltransferases.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5NR9
 
 | | A Native Ternary Complex of Alpha-1,3-Galactosyltransferase (alpha-3GalT) Supports a Conserved Reaction Mechanism for Retaining Glycosyltransferases - Unliganded alpha-3GalT | | 分子名称: | GLYCEROL, N-acetyllactosaminide alpha-1,3-galactosyltransferase | | 著者 | Albesa-Jove, D, Marina, A, Sainz-Polo, M.A, Guerin, M.E. | | 登録日 | 2017-04-22 | | 公開日 | 2017-10-11 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural Snapshots of alpha-1,3-Galactosyltransferase with Native Substrates: Insight into the Catalytic Mechanism of Retaining Glycosyltransferases.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5NRB
 
 | | A Native Ternary Complex of Alpha-1,3-Galactosyltransferase (a-3GalT) Supports a Conserved Reaction Mechanism for Retaining Glycosyltransferases - alpha-3GalT in complex with Co2+, UDP-Gal and lactose - a3GalT-Co2+-UDP-Gal-LAT-1 | | 分子名称: | COBALT (II) ION, GALACTOSE-URIDINE-5'-DIPHOSPHATE, N-acetyllactosaminide alpha-1,3-galactosyltransferase, ... | | 著者 | Albesa-Jove, D, Marina, A, Sainz-Polo, M.A, Guerin, M.E. | | 登録日 | 2017-04-22 | | 公開日 | 2017-10-11 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Structural Snapshots of alpha-1,3-Galactosyltransferase with Native Substrates: Insight into the Catalytic Mechanism of Retaining Glycosyltransferases.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5NRE
 
 | | A Native Ternary Complex of Alpha-1,3-Galactosyltransferase (a3GalT) Supports a Conserved Reaction Mechanism for Retaining Glycosyltransferases - a3GalT in complex with lactose - a3GalT-LAT | | 分子名称: | N-acetyllactosaminide alpha-1,3-galactosyltransferase, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Albesa-Jove, D, Marina, A, Sainz-Polo, M.A, Guerin, M.E. | | 登録日 | 2017-04-22 | | 公開日 | 2017-10-11 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Structural Snapshots of alpha-1,3-Galactosyltransferase with Native Substrates: Insight into the Catalytic Mechanism of Retaining Glycosyltransferases.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
