3EEO
 
 | |
3UB5
 
 | |
3U4L
 
 | |
8F3C
 
 | | Cryo-EM consensus structure of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand | | 分子名称: | DNA (38-MER), DNA (39-MER), DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | 登録日 | 2022-11-09 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8G2W
 
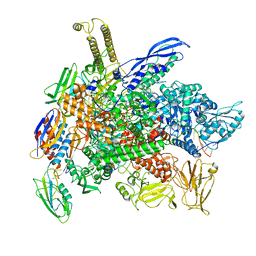 | | Cryo-EM structure of 3DVA component 2 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand | | 分子名称: | DNA (31-MER), DNA (39-MER), DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | 登録日 | 2023-02-06 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8G00
 
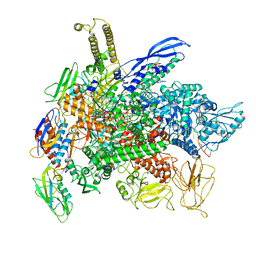 | | Cryo-EM structure of 3DVA component 0 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand | | 分子名称: | DNA (31-MER), DNA (39-mer), DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | 登録日 | 2023-01-31 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8G1S
 
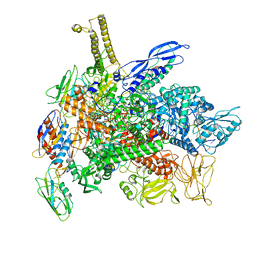 | | Cryo-EM structure of 3DVA component 1 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand | | 分子名称: | DNA (31-MER), DNA (39-MER), DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | 登録日 | 2023-02-02 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8G4W
 
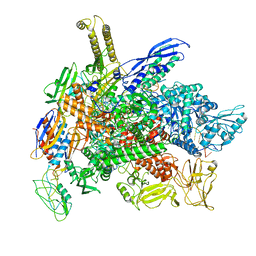 | | Cryo-EM consensus structure of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase plus preQ1 ligand | | 分子名称: | 7-DEAZA-7-AMINOMETHYL-GUANINE, DNA (31-MER), DNA (39-mer), ... | | 著者 | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | 登録日 | 2023-02-10 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8G8Z
 
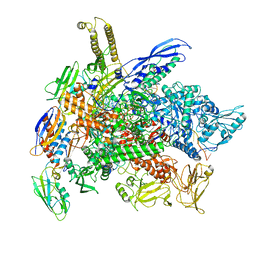 | | Cryo-EM structure of 3DVA component 1 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase plus preQ1 ligand | | 分子名称: | 7-DEAZA-7-AMINOMETHYL-GUANINE, DNA (31-MER), DNA (39-MER), ... | | 著者 | Porta, J.C, Ohi, M.D, Walter, N.G, Frank, A.T, Deb, I, Meze, K. | | 登録日 | 2023-02-20 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8G7E
 
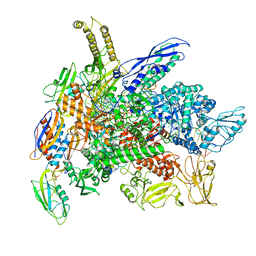 | | Cryo-EM structure of 3DVA component 0 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase plus preQ1 ligand | | 分子名称: | 7-DEAZA-7-AMINOMETHYL-GUANINE, DNA (31-MER), DNA (39-mer), ... | | 著者 | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | 登録日 | 2023-02-16 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5UHY
 
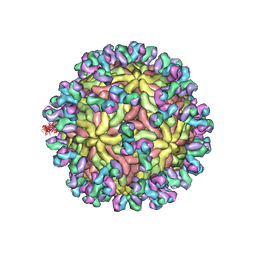 | | A Human Antibody Against Zika Virus Crosslinks the E Protein to Prevent Infection | | 分子名称: | ZV67 Fab chain 1, ZV67 Fab chain 2, envelope protein | | 著者 | Hasan, S.S, Miller, A, Sapparapu, G, Fernandez, E, Klose, T, Long, F, Fokine, A, Porta, J.C, Jiang, W, Diamond, M.S, Crowe Jr, J.E, Kuhn, R.J, Rossmann, M.G. | | 登録日 | 2017-01-12 | | 公開日 | 2017-03-29 | | 最終更新日 | 2019-11-27 | | 実験手法 | ELECTRON MICROSCOPY (6.2 Å) | | 主引用文献 | A human antibody against Zika virus crosslinks the E protein to prevent infection.
Nat Commun, 8, 2017
|
|
7SC0
 
 | |
2I5D
 
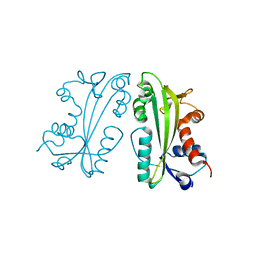 | | Crystal Structure of Human Inosine Triphosphate Pyrophosphatase | | 分子名称: | inosine triphosphate pyrophosphohydrolase | | 著者 | Porta, J.C, Kozmin, S.G, Pavlov, Y.I, Borgstahl, G.E.O. | | 登録日 | 2006-08-24 | | 公開日 | 2006-09-05 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Structure of the orthorhombic form of human inosine triphosphate pyrophosphatase.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2NYB
 
 | |
3K9S
 
 | |
