3WIQ
 
 | | Crystal structure of kojibiose phosphorylase complexed with kojibiose | | 分子名称: | Kojibiose phosphorylase, SULFATE ION, alpha-D-glucopyranose-(1-2)-beta-D-glucopyranose | | 著者 | Okada, S, Yamamoto, T, Watanabe, H, Nishimoto, T, Chaen, H, Fukuda, S, Wakagi, T, Fushinobu, S. | | 登録日 | 2013-09-24 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural and mutational analysis of substrate recognition in kojibiose phosphorylase
Febs J., 281, 2014
|
|
3WIR
 
 | | Crystal structure of kojibiose phosphorylase complexed with glucose | | 分子名称: | GLYCEROL, Kojibiose phosphorylase, PHOSPHATE ION, ... | | 著者 | Okada, S, Yamamoto, T, Watanabe, H, Nishimoto, T, Chaen, H, Fukuda, S, Wakagi, T, Fushinobu, S. | | 登録日 | 2013-09-24 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural and mutational analysis of substrate recognition in kojibiose phosphorylase
Febs J., 281, 2014
|
|
3WQH
 
 | | Crystal Structure of human DPP-IV in complex with Anagliptin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, N-[2-({2-[(2S)-2-cyanopyrrolidin-1-yl]-2-oxoethyl}amino)-2-methylpropyl]-2-methylpyrazolo[1,5-a]pyrimidine-6-carboxamide | | 著者 | Watanabe, Y.S, Okada, S, Motoyama, T, Takahashi, R, Adachi, H, Oka, M. | | 登録日 | 2014-01-27 | | 公開日 | 2015-07-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Anagliptin, a potent dipeptidyl peptidase IV inhibitor: its single-crystal structure and enzyme interactions.
J Enzyme Inhib Med Chem, 30, 2015
|
|
7YOW
 
 | |
8I5I
 
 | |
8I5H
 
 | |
7WN2
 
 | |
7WNB
 
 | |
1J0J
 
 | |
1J0I
 
 | |
5XM1
 
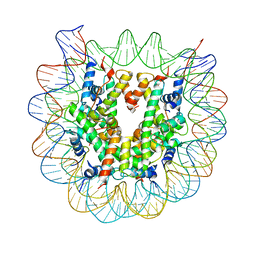 | | The mouse nucleosome structure containing H2A, H2B type3-A, H3mm7, and H4 | | 分子名称: | DNA (146-MER), Histone H2A type 1-B, Histone H2B type 3-A, ... | | 著者 | Taguchi, H, Horikoshi, N, Kurumizaka, H. | | 登録日 | 2017-05-12 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.45 Å) | | 主引用文献 | Histone H3.3 sub-variant H3mm7 is required for normal skeletal muscle regeneration.
Nat Commun, 9, 2018
|
|
1J0H
 
 | |
5XM0
 
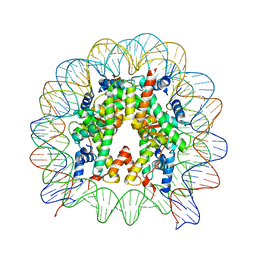 | | The mouse nucleosome structure containing H2A, H2B type3-A, H3.3, and H4 | | 分子名称: | DNA (146-MER), Histone H2A type 1-B, Histone H2B type 3-A, ... | | 著者 | Taguchi, H, Horikoshi, N, Kurumizaka, H. | | 登録日 | 2017-05-12 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.874 Å) | | 主引用文献 | Histone H3.3 sub-variant H3mm7 is required for normal skeletal muscle regeneration.
Nat Commun, 9, 2018
|
|
1J0K
 
 | |
1C58
 
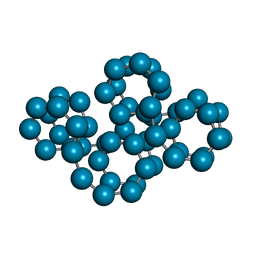 | | CRYSTAL STRUCTURE OF CYCLOAMYLOSE 26 | | 分子名称: | Cyclohexacosakis-(1-4)-(alpha-D-glucopyranose) | | 著者 | Gessler, K, Saenger, W, Nimz, O. | | 登録日 | 1999-11-04 | | 公開日 | 1999-11-10 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (0.99 Å) | | 主引用文献 | V-Amylose at atomic resolution: X-ray structure of a cycloamylose with 26 glucose residues (cyclomaltohexaicosaose).
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1BWV
 
 | | Activated Ribulose 1,5-Bisphosphate Carboxylase/Oxygenase (RUBISCO) Complexed with the Reaction Intermediate Analogue 2-Carboxyarabinitol 1,5-Bisphosphate | | 分子名称: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, PROTEIN (RIBULOSE BISPHOSPHATE CARBOXYLASE) | | 著者 | Sugawara, H, Yamamoto, H, Shibata, N, Inoue, T, Miyake, C, Yokota, A, Kai, Y. | | 登録日 | 1998-09-29 | | 公開日 | 1999-09-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of carboxylase reaction-oriented ribulose 1, 5-bisphosphate carboxylase/oxygenase from a thermophilic red alga, Galdieria partita.
J.Biol.Chem., 274, 1999
|
|
