1W7A
 
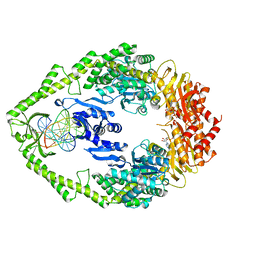 | | ATP bound MutS | | 分子名称: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP*CP*CP*TP* AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP *GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP*GP*GP*CP*AP*GP* CP*T)-3', ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Lamers, M.H, Georgijevic, D, Lebbink, J, Winterwerp, H.H.K, Agianian, B, de Wind, N, Sixma, T.K. | | 登録日 | 2004-08-31 | | 公開日 | 2004-09-10 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | ATP Increases the Affinity between Muts ATPase Domains: Implications for ATP Hydrolysis and Conformational Changes
J.Biol.Chem., 279, 2004
|
|
1NG9
 
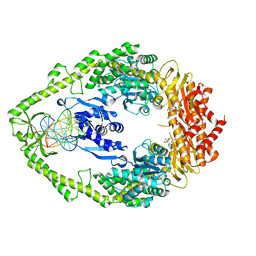 | | E.coli MutS R697A: an ATPase-asymmetry mutant | | 分子名称: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP*CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP*GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP*GP*GP*CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Lamers, M.H, Winterwerp, H.H.K, Sixma, T.K. | | 登録日 | 2002-12-17 | | 公開日 | 2003-02-11 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The alternating ATPase domains of MutS control DNA mismatch repair
Embo J., 22, 2003
|
|
2HQA
 
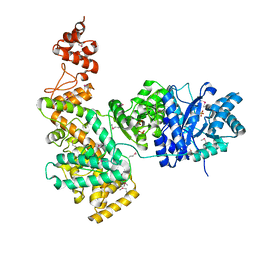 | | Crystal structure of the catalytic alpha subunit of E. Coli replicative DNA polymerase III | | 分子名称: | DNA polymerase III alpha subunit, PHOSPHATE ION | | 著者 | Lamers, M.H, Georgescu, R.E, Lee, S.G, O'Donnell, M, Kuriyan, J. | | 登録日 | 2006-07-18 | | 公開日 | 2006-09-19 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal Structure of the Catalytic alpha Subunit of E. coli Replicative DNA Polymerase III.
Cell(Cambridge,Mass.), 126, 2006
|
|
1E3M
 
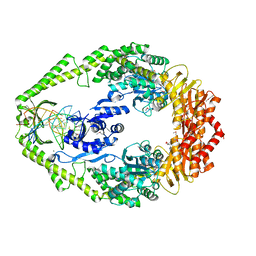 | | The crystal structure of E. coli MutS binding to DNA with a G:T mismatch | | 分子名称: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*CP*AP*CP*CP*AP* GP*TP*GP*TP*CP*AP*GP*CP*GP*TP*CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP*GP*AP*CP*AP*CP* TP*GP*GP*TP*GP*CP*TP*TP*GP*GP*CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Lamers, M.H, Perrakis, A, Enzlin, J.H, Winterwerp, H.H.K, De Wind, N, Sixma, T.K. | | 登録日 | 2000-06-19 | | 公開日 | 2000-11-01 | | 最終更新日 | 2017-07-05 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Crystal Structure of DNA Mismatch Repair Protein Muts Binding to a G X T Mismatch
Nature, 407, 2000
|
|
7OTO
 
 | | The structure of MutS bound to two molecules of AMPPNP | | 分子名称: | DNA mismatch repair protein MutS, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Lamers, M.H, Borsellini, A, Friedhoff, P, Kunetsky, V. | | 登録日 | 2021-06-10 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryogenic electron microscopy structures reveal how ATP and DNA binding in MutS coordinates sequential steps of DNA mismatch repair.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7OU4
 
 | | The structure of MutS bound to one molecule of ATP and one molecule of ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA mismatch repair protein MutS, ... | | 著者 | Lamers, M.H, Borsellini, A, Friedhoff, P, Kunetsky, V. | | 登録日 | 2021-06-11 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryogenic electron microscopy structures reveal how ATP and DNA binding in MutS coordinates sequential steps of DNA mismatch repair.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7OU0
 
 | | The structure of MutS bound to two molecules of ADP-Vanadate | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein MutS, MAGNESIUM ION, ... | | 著者 | Lamers, M.H, Borsellini, A, Friedhoff, P, Kunetsky, V. | | 登録日 | 2021-06-10 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryogenic electron microscopy structures reveal how ATP and DNA binding in MutS coordinates sequential steps of DNA mismatch repair.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7OU2
 
 | | The structure of MutS bound to two molecules of ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein MutS | | 著者 | Lamers, M.H, Borsellini, A, Friedhoff, P, Kunetsky, V. | | 登録日 | 2021-06-11 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Cryogenic electron microscopy structures reveal how ATP and DNA binding in MutS coordinates sequential steps of DNA mismatch repair.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8OOY
 
 | | Pol I bound to extended and displaced DNA section - open conformation | | 分子名称: | DNA polymerase I, Displacing Primer, Extending Primer, ... | | 著者 | Botto, M, Borsellini, A, Lamers, M.H. | | 登録日 | 2023-04-06 | | 公開日 | 2023-08-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | A four-point molecular handover during Okazaki maturation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8OO6
 
 | | Pol I bound to extended and displaced DNA section - closed conformation | | 分子名称: | DNA polymerase I, Displaced primer, Extending Primer, ... | | 著者 | Botto, M, Borsellini, A, Lamers, M.H. | | 登録日 | 2023-04-04 | | 公開日 | 2023-08-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | A four-point molecular handover during Okazaki maturation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3GT8
 
 | | Crystal structure of the inactive EGFR kinase domain in complex with AMP-PNP | | 分子名称: | Epidermal growth factor receptor, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Jura, N, Endres, N.F, Engel, K, Deindl, S, Das, R, Lamers, M.H, Wemmer, D.E, Zhang, X, Kuriyan, J. | | 登録日 | 2009-03-27 | | 公開日 | 2009-07-21 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.955 Å) | | 主引用文献 | Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment.
Cell(Cambridge,Mass.), 137, 2009
|
|
2YMB
 
 | | Structures of MITD1 | | 分子名称: | CHARGED MULTIVESICULAR BODY PROTEIN 1A, MIT DOMAIN-CONTAINING PROTEIN 1 | | 著者 | Hadders, M.A, Agromayor, M, Obita, T, Perisic, O, Caballe, A, Kloc, M, Lamers, M.H, Williams, R.L, Martin-Serrano, J. | | 登録日 | 2012-10-08 | | 公開日 | 2012-10-24 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.404 Å) | | 主引用文献 | Escrt-III Binding Protein Mitd1 is Involved in Cytokinesis and Has an Unanticipated Pld Fold that Binds Membranes.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5LEW
 
 | | DNA polymerase | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA polymerase III subunit alpha, SULFATE ION, ... | | 著者 | Banos-Mateos, S, Lang, U.F, Maslen, S.L, Skehel, J.M, Lamers, M.H. | | 登録日 | 2016-06-30 | | 公開日 | 2017-10-25 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | High-fidelity DNA replication in Mycobacterium tuberculosis relies on a trinuclear zinc center.
Nat Commun, 8, 2017
|
|
5M1S
 
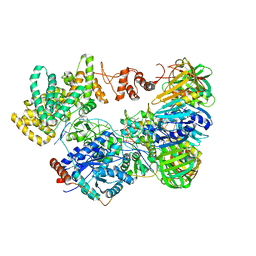 | | Cryo-EM structure of the E. coli replicative DNA polymerase-clamp-exonuclase-theta complex bound to DNA in the editing mode | | 分子名称: | DNA Primer Strand, DNA Template Strand, DNA polymerase III subunit alpha, ... | | 著者 | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | 登録日 | 2016-10-10 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (6.7 Å) | | 主引用文献 | Self-correcting mismatches during high-fidelity DNA replication.
Nat. Struct. Mol. Biol., 24, 2017
|
|
1OH5
 
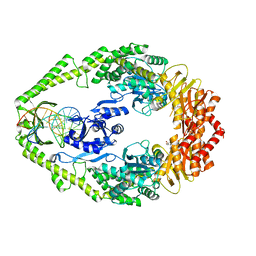 | | THE CRYSTAL STRUCTURE OF E. COLI MUTS BINDING TO DNA WITH A C:A MISMATCH | | 分子名称: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*CP*GP *CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP *CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP *GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*AP*TP*GP*GP *CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Natrajan, G, Lamers, M.H, Enzlin, J.H, Winterwerp, H.H.K, Perrakis, A, Sixma, T.K. | | 登録日 | 2003-05-23 | | 公開日 | 2003-08-08 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structures of E. Coli DNA Mismatch Repair Enzyme Muts in Complex with Different Mismatches: A Common Recognition Mode for Diverse Substrates
Nucleic Acids Res., 31, 2003
|
|
1OH6
 
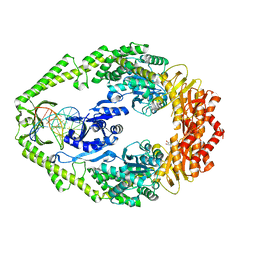 | | THE CRYSTAL STRUCTURE OF E. COLI MUTS BINDING TO DNA WITH AN A:A MISMATCH | | 分子名称: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*AP*GP *CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP* CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP *GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*AP*TP*GP*GP* CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Natrajan, G, Lamers, M.H, Enzlin, J.H, Winterwerp, H.H.K, Perrakis, A, Sixma, T.K. | | 登録日 | 2003-05-23 | | 公開日 | 2003-08-08 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structures of E. Coli DNA Mismatch Repair Enzyme Muts in Complex with Different Mismatches: A Common Recognition Mode for Diverse Substrates
Nucleic Acids Res., 31, 2003
|
|
1OH8
 
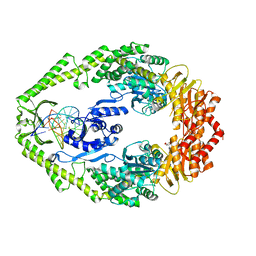 | | THE CRYSTAL STRUCTURE OF E. COLI MUTS BINDING TO DNA WITH AN UNPAIRED THYMIDINE | | 分子名称: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP*CP*CP* TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP *GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*CP*TP*TP*GP*GP*CP* AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Natrajan, G, Lamers, M.H, Enzlin, J.H, Winterwerp, H.H.K, Perrakis, A, Sixma, T.K. | | 登録日 | 2003-05-23 | | 公開日 | 2003-08-08 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structures of E. Coli DNA Mismatch Repair Enzyme Muts in Complex with Different Mismatches: A Common Recognition Mode for Diverse Substrates
Nucleic Acids Res., 31, 2003
|
|
1OH7
 
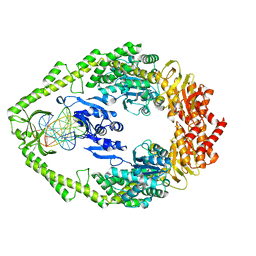 | | THE CRYSTAL STRUCTURE OF E. COLI MUTS BINDING TO DNA WITH A G:G MISMATCH | | 分子名称: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP *CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP *GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*GP*TP*GP*GP *CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Natrajan, G, Lamers, M.H, Enzlin, J.H, Winterwerp, H.H.K, Perrakis, A, Sixma, T.K. | | 登録日 | 2003-05-23 | | 公開日 | 2003-08-08 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures of E. Coli DNA Mismatch Repair Enzyme Muts in Complex with Different Mismatches: A Common Recognition Mode for Diverse Substrates
Nucleic Acids Res., 31, 2003
|
|
7P8V
 
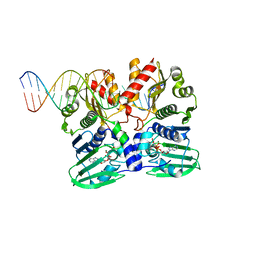 | |
4JOM
 
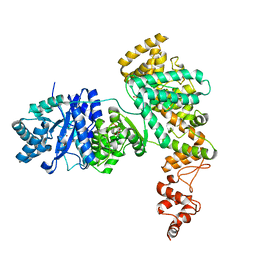 | | Structure of E. coli Pol III 3mPHP mutant | | 分子名称: | DNA polymerase III subunit alpha, GLYCEROL, PHOSPHATE ION, ... | | 著者 | Barros, T, Guenther, J, Kelch, B, Anaya, J, Prabhakar, A, O'Donnell, M, Kuriyan, J, Lamers, M.H. | | 登録日 | 2013-03-18 | | 公開日 | 2013-05-29 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | A structural role for the PHP domain in E. coli DNA polymerase III.
Bmc Struct.Biol., 13, 2013
|
|
5FKV
 
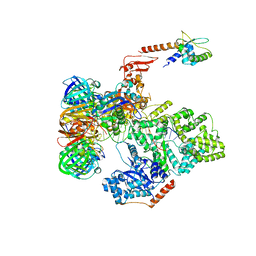 | | cryo-EM structure of the E. coli replicative DNA polymerase complex bound to DNA (DNA polymerase III alpha, beta, epsilon, tau complex) | | 分子名称: | DNA POLYMERASE III BETA, DNA POLYMERASE III EPSILON, DNA POLYMERASE III SUBUNIT ALPHA, ... | | 著者 | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | 登録日 | 2015-10-20 | | 公開日 | 2015-11-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (8.04 Å) | | 主引用文献 | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
5FKW
 
 | | cryo-EM structure of the E. coli replicative DNA polymerase complex bound to DNA (DNA polymerase III alpha, beta, epsilon) | | 分子名称: | DNA POLYMERASE III ALPHA, DNA POLYMERASE III BETA, DNA POLYMERASE III EPSILON, ... | | 著者 | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | 登録日 | 2015-10-20 | | 公開日 | 2015-11-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7.3 Å) | | 主引用文献 | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
5FKU
 
 | | cryo-EM structure of the E. coli replicative DNA polymerase complex in DNA free state (DNA polymerase III alpha, beta, epsilon, tau complex) | | 分子名称: | DNA POLYMERASE III SUBUNIT ALPHA, DNA POLYMERASE III SUBUNIT BETA, DNA POLYMERASE III SUBUNIT EPSILON, ... | | 著者 | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | 登録日 | 2015-10-20 | | 公開日 | 2015-11-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (8.34 Å) | | 主引用文献 | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
3ET6
 
 | | The crystal structure of the catalytic domain of a eukaryotic guanylate cyclase | | 分子名称: | PHOSPHATE ION, Soluble guanylyl cyclase beta | | 著者 | Winger, J.A, Derbyshire, E.R, Lamers, M.H, Marletta, M.A, Kuriyan, J. | | 登録日 | 2008-10-07 | | 公開日 | 2008-10-14 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | The crystal structure of the catalytic domain of a eukaryotic guanylate cyclase.
Bmc Struct.Biol., 8, 2008
|
|
7AI7
 
 | | MutS in Intermediate state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*CP*TP*TP*AP*GP*CP*TP*TP*AP*GP*GP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*CP*CP*TP*AP*AP*CP*TP*AP*AP*G)-3'), ... | | 著者 | Fernandez-Leiro, R, Bhairosing-Kok, D, Sixma, T.K, Lamers, M.H. | | 登録日 | 2020-09-26 | | 公開日 | 2021-03-31 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | The selection process of licensing a DNA mismatch for repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
