2F3I
 
 | |
2LIZ
 
 | |
8QEW
 
 | |
9M6Y
 
 | |
3EBN
 
 | | A Special Dimerization of SARS-CoV Main Protease C-Terminal Domain Due to Domain-swapping | | 分子名称: | Replicase polyprotein 1ab | | 著者 | Zhong, N, Zhang, S, Xue, F, Kang, X, Lou, Z, Xia, B. | | 登録日 | 2008-08-28 | | 公開日 | 2009-05-19 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | C-terminal domain of SARS-CoV main protease can form a 3D domain-swapped dimer
PROTEIN SCI., 18, 2009
|
|
5HRC
 
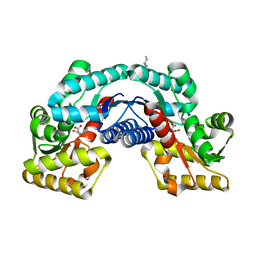 | | Crystal structure of an aspartate/glutamate racemase in complex with L-aspartate | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ASPARTIC ACID, aspartate/glutamate racemase | | 著者 | Liu, X, Gao, F, Ma, Y, Liu, S, Cui, Y, Yuan, Z, Kang, X. | | 登録日 | 2016-01-23 | | 公開日 | 2016-04-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.765 Å) | | 主引用文献 | Crystal structure and molecular mechanism of an aspartate/glutamate racemase from Escherichia coli O157
Febs Lett., 590, 2016
|
|
5HRA
 
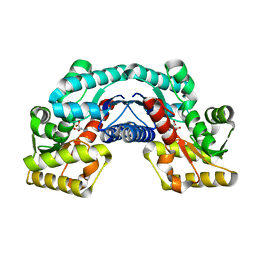 | | Crystal structure of an aspartate/glutamate racemase in complex with D-aspartate | | 分子名称: | D-ASPARTIC ACID, aspartate/glutamate racemase | | 著者 | Liu, X, Gao, F, Ma, Y, Liu, S, Cui, Y, Yuan, Z, Kang, X. | | 登録日 | 2016-01-23 | | 公開日 | 2016-04-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.597 Å) | | 主引用文献 | Crystal structure and molecular mechanism of an aspartate/glutamate racemase from Escherichia coli O157
Febs Lett., 590, 2016
|
|
5HQT
 
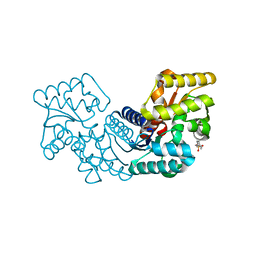 | | Crystal structure of an aspartate/glutamate racemase from Escherichia coli O157 | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, aspartate/glutamate racemase | | 著者 | Liu, X, Gao, F, Ma, Y, Liu, S, Cui, Y, Yuan, Z, Kang, X. | | 登録日 | 2016-01-22 | | 公開日 | 2016-04-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.598 Å) | | 主引用文献 | Crystal structure and molecular mechanism of an aspartate/glutamate racemase from Escherichia coli O157
Febs Lett., 590, 2016
|
|
8YKE
 
 | |
9AVK
 
 | | Structure of long Rib domain from Limosilactobacillus reuteri | | 分子名称: | SODIUM ION, THIOCYANATE ION, YSIRK signal domain/LPXTG anchor domain surface protein | | 著者 | Xue, Y, Kang, X. | | 登録日 | 2024-03-04 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Crystal structure of the long Rib domain of the LPXTG-anchored surface protein from Limosilactobacillus reuteri.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
8YK7
 
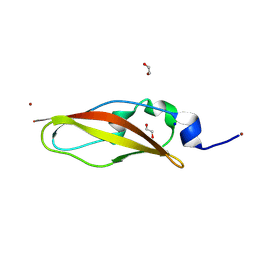 | | Structure of Rib domain from surface adhesin of Limosilactobacillus reuteri | | 分子名称: | 1,2-ETHANEDIOL, COPPER (II) ION, SODIUM ION, ... | | 著者 | Xue, Y, Kang, X. | | 登録日 | 2024-03-04 | | 公開日 | 2024-09-11 | | 最終更新日 | 2024-09-18 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Crystal structure of the Rib domain of the cell-wall-anchored surface protein from Limosilactobacillus reuteri.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
3WDO
 
 | | Structure of E. coli YajR transporter | | 分子名称: | MFS Transporter | | 著者 | Jiang, D. | | 登録日 | 2013-06-19 | | 公開日 | 2013-08-07 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Structure of the YajR transporter suggests a transport mechanism based on the conserved motif A
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3IWM
 
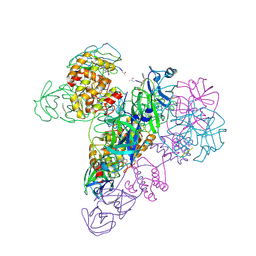 | | The octameric SARS-CoV main protease | | 分子名称: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | 著者 | Zhong, N, Zhang, S, Xue, F, Lou, Z, Rao, Z, Xia, B. | | 登録日 | 2009-09-02 | | 公開日 | 2010-07-21 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Three-dimensional domain swapping as a mechanism to lock the active conformation in a super-active octamer of SARS-CoV main protease
Protein Cell, 1, 2010
|
|
7K6M
 
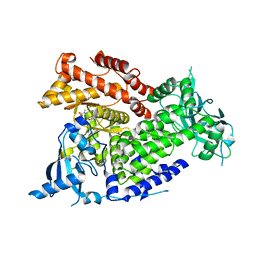 | | Crystal structure of PI3Kalpha selective Inhibitor PF-06843195 | | 分子名称: | 2,2-difluoroethyl (3S)-3-{[2'-amino-5-fluoro-2-(morpholin-4-yl)[4,5'-bipyrimidin]-6-yl]amino}-3-(hydroxymethyl)pyrrolidine-1-carboxylate, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | 著者 | Chen, P, Brooun, A, Deng, Y.L, Grodsky, N, Kaiser, S.E. | | 登録日 | 2020-09-21 | | 公開日 | 2021-01-06 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.413 Å) | | 主引用文献 | Structure-Based Drug Design and Synthesis of PI3K alpha-Selective Inhibitor (PF-06843195).
J.Med.Chem., 64, 2021
|
|
2JSO
 
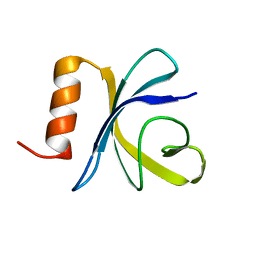 | | Antimicrobial resistance protein | | 分子名称: | Polymyxin resistance protein pmrD | | 著者 | Jin, C, Fu, W. | | 登録日 | 2007-07-10 | | 公開日 | 2007-09-04 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | First structure of the polymyxin resistance proteins.
Biochem.Biophys.Res.Commun., 361, 2007
|
|
4TZ7
 
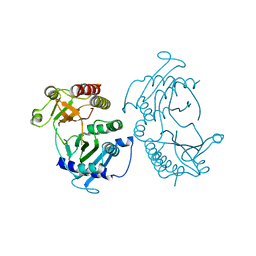 | | Crystal structure of type I phosphatidylinositol 4-phosphate 5-kinase alpha from Zebrafish | | 分子名称: | Phosphatidylinositol-4-phosphate 5-kinase, type I, alpha | | 著者 | Hu, J, Qin, Y, Wang, J, Li, L, Wu, D, Ha, Y. | | 登録日 | 2014-07-09 | | 公開日 | 2015-09-02 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.31 Å) | | 主引用文献 | Resolution of structure of PIP5K1A reveals molecular mechanism for its regulation by dimerization and dishevelled.
Nat Commun, 6, 2015
|
|
7WKX
 
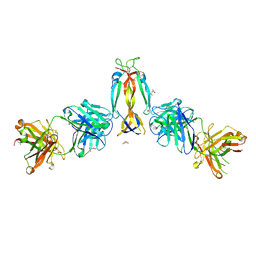 | | IL-17A in complex with the humanized antibody HB0017 | | 分子名称: | ACETIC ACID, Heavy chain of HB0017 Fab, Interleukin-17A, ... | | 著者 | Xu, J, Zhu, X, He, Y. | | 登録日 | 2022-01-12 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Structural and functional insights into a novel pre-clinical-stage antibody targeting IL-17A for treatment of autoimmune diseases.
Int.J.Biol.Macromol., 202, 2022
|
|
7WRH
 
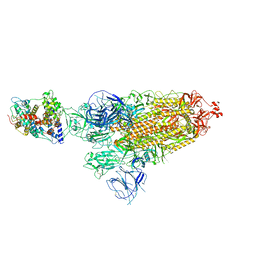 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.1 spike protein in complex with mouse ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Han, P, Xie, Y, Qi, J. | | 登録日 | 2022-01-26 | | 公開日 | 2023-02-01 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (2.66 Å) | | 主引用文献 | Broader-species receptor binding and structural bases of Omicron SARS-CoV-2 to both mouse and palm-civet ACE2s.
Cell Discov, 8, 2022
|
|
7WRI
 
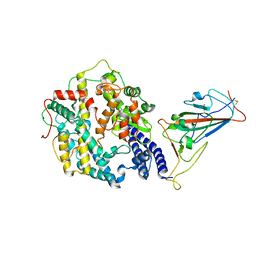 | | Cryo-EM structure of SARS-CoV-2 Omicron spike receptor-binding domain in complex with mouse ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein, ... | | 著者 | Han, P, Xie, Y, Qi, J. | | 登録日 | 2022-01-26 | | 公開日 | 2022-06-08 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | Broader-species receptor binding and structural bases of Omicron SARS-CoV-2 to both mouse and palm-civet ACE2s.
Cell Discov, 8, 2022
|
|
7WSK
 
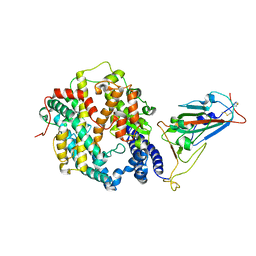 | | Crystal structure of SARS-CoV-2 Omicron spike receptor-binding domain in complex with civet ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1, ... | | 著者 | Huang, B, Han, P, Qi, J. | | 登録日 | 2022-01-29 | | 公開日 | 2022-06-08 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Broader-species receptor binding and structural bases of Omicron SARS-CoV-2 to both mouse and palm-civet ACE2s.
Cell Discov, 8, 2022
|
|
5HXD
 
 | | Crystal structure of murein-tripeptide amidase MpaA from Escherichia coli O157 | | 分子名称: | CACODYLATE ION, Protein MpaA, ZINC ION | | 著者 | Ma, Y, Bai, G, Zhang, X, Zhao, J, Yuan, Z, Kang, X, Li, Z, Mu, S, Liu, X. | | 登録日 | 2016-01-30 | | 公開日 | 2017-02-08 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal Structure of Murein-Tripeptide Amidase MpaA from Escherichia coli O157 at 2.6 angstrom Resolution
Protein Pept.Lett., 24, 2017
|
|
8XZB
 
 | | The structure of fox ACE2 and SARS-CoV RBD complex | | 分子名称: | Angiotensin-converting enzyme, Spike protein S1, ZINC ION | | 著者 | sun, J.Q. | | 登録日 | 2024-01-21 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.12 Å) | | 主引用文献 | The binding and structural basis of fox ACE2 to RBDs from different sarbecoviruses.
Virol Sin, 39, 2024
|
|
8XYZ
 
 | | The structure of fox ACE2 and PT RBD complex | | 分子名称: | Angiotensin-converting enzyme, Signal peptide, Spike protein S1, ... | | 著者 | sun, J.Q. | | 登録日 | 2024-01-20 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (2.96 Å) | | 主引用文献 | The binding and structural basis of fox ACE2 to RBDs from different sarbecoviruses.
Virol Sin, 39, 2024
|
|
8XZD
 
 | |
2K7X
 
 | |
