4XQD
 
 | | X-ray structure analysis of xylanase-WT at pH4.0 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endo-1,4-beta-xylanase 2, IODIDE ION | | 著者 | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | 登録日 | 2015-01-19 | | 公開日 | 2015-09-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XQW
 
 | | X-ray structure analysis of xylanase-N44E with MES at pH6.0 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase 2, IODIDE ION | | 著者 | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | 登録日 | 2015-01-20 | | 公開日 | 2015-09-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XPV
 
 | | Neutron and X-ray structure analysis of xylanase: N44D at pH6 | | 分子名称: | Endo-1,4-beta-xylanase 2, IODIDE ION | | 著者 | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | 登録日 | 2015-01-18 | | 公開日 | 2015-09-30 | | 最終更新日 | 2023-09-27 | | 実験手法 | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | 主引用文献 | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XQ4
 
 | | X-ray structure analysis of xylanase - N44D | | 分子名称: | Endo-1,4-beta-xylanase 2, IODIDE ION | | 著者 | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | 登録日 | 2015-01-19 | | 公開日 | 2015-09-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1QWG
 
 | | Crystal structure of Methanococcus jannaschii phosphosulfolactate synthase | | 分子名称: | (2R)-phospho-3-sulfolactate synthase, SULFATE ION | | 著者 | Wise, E.L, Graham, D.E, White, R.H, Rayment, I. | | 登録日 | 2003-09-02 | | 公開日 | 2003-12-09 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The structural determination of phosphosulfolactate synthase from Methanococcus jannaschii at 1.7-A resolution: an enolase that is not an enolase
J.Biol.Chem., 278, 2003
|
|
1MT1
 
 | | The Crystal Structure of Pyruvoyl-dependent Arginine Decarboxylase from Methanococcus jannaschii | | 分子名称: | AGMATINE, PYRUVOYL-DEPENDENT ARGININE DECARBOXYLASE ALPHA CHAIN, PYRUVOYL-DEPENDENT ARGININE DECARBOXYLASE BETA CHAIN | | 著者 | Tolbert, W.D, Graham, D.E, White, R.H, Ealick, S.E. | | 登録日 | 2002-09-20 | | 公開日 | 2003-03-25 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Pyruvoyl-Dependent Arginine Decarboxylase from Methanococcus jannaschii:
Crystal Structures of the Self-Cleaved and S53A Proenzyme Forms
Structure, 11, 2003
|
|
1N2M
 
 | | The S53A Proenzyme Structure of Methanococcus jannaschii. | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, Pyruvoyl-dependent arginine decarboxylase | | 著者 | Tolbert, W.D, Graham, D.E, White, R.H, Ealick, S.E. | | 登録日 | 2002-10-23 | | 公開日 | 2003-03-25 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Pyruvoyl-Dependent Arginine Decarboxylase from Methanococcus jannaschii:
Crystal Structures of the Self-Cleaved and S53A Proenzyme Forms
Structure, 11, 2003
|
|
1N13
 
 | | The Crystal Structure of Pyruvoyl-dependent Arginine Decarboxylase from Methanococcus jannashii | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, AGMATINE, Pyruvoyl-dependent arginine decarboxylase alpha chain, ... | | 著者 | Tolbert, W.D, Graham, D.E, White, R.H, Ealick, S.E. | | 登録日 | 2002-10-16 | | 公開日 | 2003-03-25 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Pyruvoyl-Dependent Arginine Decarboxylase from Methanococcus jannaschii:
Crystal Structures of the Self-Cleaved and S53A Proenzyme Forms
Structure, 11, 2003
|
|
5ZO0
 
 | |
5ZKZ
 
 | |
5ZII
 
 | |
5ZIW
 
 | |
2PKP
 
 | | Crystal structure of 3-isopropylmalate dehydratase (leuD)from Methhanocaldococcus Jannaschii DSM2661 (MJ1271) | | 分子名称: | DI(HYDROXYETHYL)ETHER, Homoaconitase small subunit, ZINC ION | | 著者 | Jeyakanthan, J, Gayathri, D.R, Velmurugan, D, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-04-18 | | 公開日 | 2008-04-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Substrate specificity determinants of the methanogen homoaconitase enzyme: structure and function of the small subunit
Biochemistry, 49, 2010
|
|
4QE5
 
 | |
4QE4
 
 | |
4QEH
 
 | |
4QDP
 
 | | Joint X-ray and neutron structure of Streptomyces rubiginosus D-xylose isomerase in complex with two Cd2+ ions and cyclic beta-L-arabinose | | 分子名称: | CADMIUM ION, Xylose isomerase, beta-L-arabinopyranose | | 著者 | Kovalevsky, A.Y, Langan, P. | | 登録日 | 2014-05-14 | | 公開日 | 2014-09-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | 主引用文献 | L-Arabinose Binding, Isomerization, and Epimerization by D-Xylose Isomerase: X-Ray/Neutron Crystallographic and Molecular Simulation Study.
Structure, 22, 2014
|
|
4QE1
 
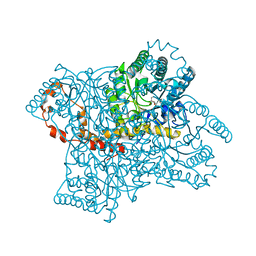 | |
4S2G
 
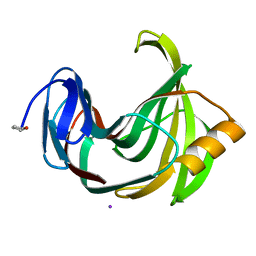 | | Joint X-ray/neutron structure of Trichoderma reesei xylanase II at pH 5.8 | | 分子名称: | Endo-1,4-beta-xylanase 2, IODIDE ION | | 著者 | Kovalevsky, A, Wan, Q, Langan, P. | | 登録日 | 2015-01-20 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-11-20 | | 実験手法 | NEUTRON DIFFRACTION (1.6 Å), X-RAY DIFFRACTION | | 主引用文献 | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4S2D
 
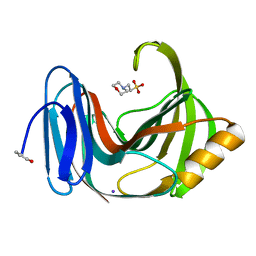 | | Joint X-ray/neutron structure of Trichoderma reesei xylanase II in complex with MES at pH 5.7 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase 2, IODIDE ION | | 著者 | Kovalevsky, A.Y, Wan, Q, Langan, P. | | 登録日 | 2015-01-20 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-11-27 | | 実験手法 | NEUTRON DIFFRACTION (1.6 Å), X-RAY DIFFRACTION | | 主引用文献 | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4S2F
 
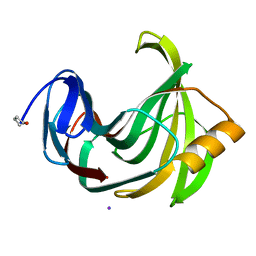 | | Joint X-ray/neutron structure of Trichoderma reesei xylanase II at pH 4.4 | | 分子名称: | Endo-1,4-beta-xylanase 2, IODIDE ION | | 著者 | Kovalevsky, A, Wan, Q, Langan, P. | | 登録日 | 2015-01-20 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-11-20 | | 実験手法 | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | 主引用文献 | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4S2H
 
 | | Joint X-ray/neutron structure of Trichoderma reesei xylanase II at pH 8.5 | | 分子名称: | Endo-1,4-beta-xylanase 2, IODIDE ION | | 著者 | Kovalevsky, A, Wan, Q, Langan, P. | | 登録日 | 2015-01-20 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-11-20 | | 実験手法 | NEUTRON DIFFRACTION (1.6 Å), X-RAY DIFFRACTION | | 主引用文献 | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4QEE
 
 | |
4QDW
 
 | | Joint X-ray and neutron structure of Streptomyces rubiginosus D-xylose isomerase in complex with two Ni2+ ions and linear L-arabinose | | 分子名称: | L-arabinose, NICKEL (II) ION, Xylose isomerase | | 著者 | Kovalevsky, A.Y, Langan, P. | | 登録日 | 2014-05-14 | | 公開日 | 2014-09-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | 主引用文献 | L-Arabinose Binding, Isomerization, and Epimerization by D-Xylose Isomerase: X-Ray/Neutron Crystallographic and Molecular Simulation Study.
Structure, 22, 2014
|
|
