2MBS
 
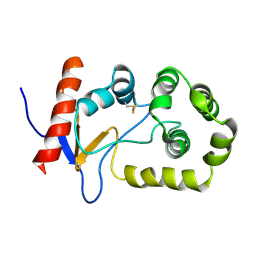 | | NMR solution structure of oxidized KpDsbA | | 分子名称: | Thiol:disulfide interchange protein | | 著者 | Kurth, F, Rimmer, K, Premkumar, L, Mohanty, B, Duprez, W, Halili, M.A, Shouldice, S.R, Heras, B, Fairlie, D.P, Scanlon, M.J, Martin, J.L. | | 登録日 | 2013-08-03 | | 公開日 | 2013-12-11 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comparative Sequence, Structure and Redox Analyses of Klebsiella pneumoniae DsbA Show That Anti-Virulence Target DsbA Enzymes Fall into Distinct Classes.
Plos One, 8, 2013
|
|
4TKY
 
 | |
4MCU
 
 | |
4K6X
 
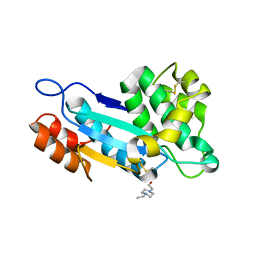 | | Crystal structure of disulfide oxidoreductase from Mycobacterium tuberculosis | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Disulfide oxidoreductase | | 著者 | Premkumar, L, Martin, J.L. | | 登録日 | 2013-04-16 | | 公開日 | 2013-10-02 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.972 Å) | | 主引用文献 | Rv2969c, essential for optimal growth in Mycobacterium tuberculosis, is a DsbA-like enzyme that interacts with VKOR-derived peptides and has atypical features of DsbA-like disulfide oxidases.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
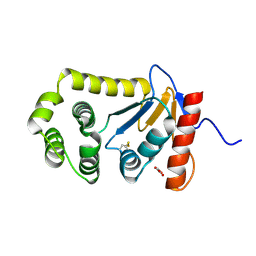
4OCE
 
 | |
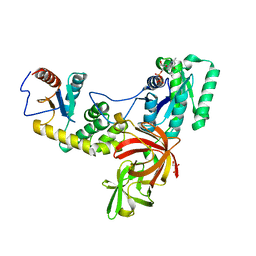
4P3Y
 
 | |
4OCF
 
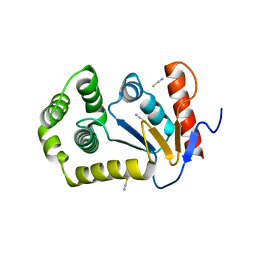 | |
4OD7
 
 | |
