6BQE
 
 | |
8HQQ
 
 | |
8HQR
 
 | |
8KD0
 
 | |
5JOS
 
 | |
5KKW
 
 | |
5HPQ
 
 | |
8WCH
 
 | |
5WJP
 
 | |
4ZV1
 
 | |
4ZV2
 
 | |
5T0W
 
 | |
5TUJ
 
 | |
6WUP
 
 | |
4N1C
 
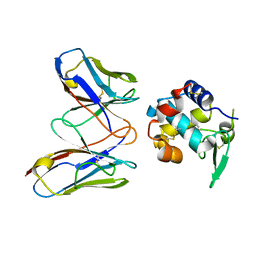 | | Structural evidence for antigen receptor evolution | | 分子名称: | Lysozyme C, immunoglobulin variable light chain domain | | 著者 | Langley, D.B, Rouet, R, Roome, B, Stock, D, Christ, D. | | 登録日 | 2013-10-03 | | 公開日 | 2014-10-29 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural reconstruction of protein ancestry.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4N1E
 
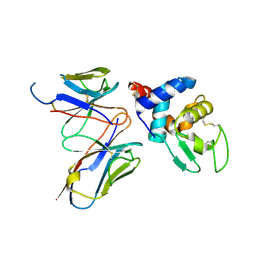 | | Structural evidence for antigen receptor evolution | | 分子名称: | Lysozyme C, immunoglobulin variable light chain domain | | 著者 | Langley, D.B, Rouet, R, Stock, D, Christ, D. | | 登録日 | 2013-10-04 | | 公開日 | 2014-10-29 | | 最終更新日 | 2018-04-18 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Structural reconstruction of protein ancestry.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7KWW
 
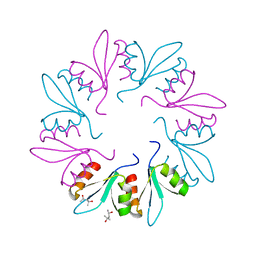 | | X-ray Crystal Structure of PlyCB Mutant K59H | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, PlyCB | | 著者 | Williams, D.E, Broendum, S.S, Hayes, B.K, Drinkwater, N, McGowan, S. | | 登録日 | 2020-12-02 | | 公開日 | 2021-04-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | High avidity drives the interaction between the streptococcal C1 phage endolysin, PlyC, with the cell surface carbohydrates of Group A Streptococcus.
Mol.Microbiol., 116, 2021
|
|
7KWT
 
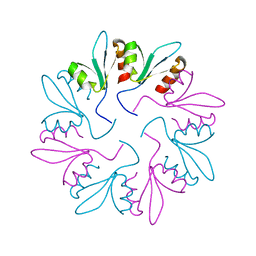 | | X-ray Crystal Structure of PlyCB Mutant Y28H | | 分子名称: | PlyCB | | 著者 | Williams, D.E, Broendum, S.S, Hayes, B.K, Drinkwater, N, McGowan, S. | | 登録日 | 2020-12-02 | | 公開日 | 2021-04-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | High avidity drives the interaction between the streptococcal C1 phage endolysin, PlyC, with the cell surface carbohydrates of Group A Streptococcus.
Mol.Microbiol., 116, 2021
|
|
7KWY
 
 | | X-ray Crystal Structure of PlyCB Mutant R66K | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, PlyCB | | 著者 | Williams, D.E, Broendum, S.S, Hayes, B.K, Drinkwater, N, McGowan, S. | | 登録日 | 2020-12-02 | | 公開日 | 2021-04-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | High avidity drives the interaction between the streptococcal C1 phage endolysin, PlyC, with the cell surface carbohydrates of Group A Streptococcus.
Mol.Microbiol., 116, 2021
|
|
5V3B
 
 | |
5V3P
 
 | |
