4YVM
 
 | |
2ZL7
 
 | | Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus | | 分子名称: | 58 kd capsid protein, ACETATE ION, CALCIUM ION, ... | | 著者 | Choi, J.M, Huston, A.M, Estes, M.K, Prasad, B.V.V. | | 登録日 | 2008-04-02 | | 公開日 | 2008-07-22 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2ZL5
 
 | | Atomic resolution structural characterization of recognition of histo-blood group antigen by Norwalk virus | | 分子名称: | 58 kd capsid protein, ACETATE ION, CALCIUM ION, ... | | 著者 | Choi, J.M, Huston, A.M, Estes, M.K, Prasad, B.V.V. | | 登録日 | 2008-04-02 | | 公開日 | 2008-07-22 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2ZL6
 
 | | Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus | | 分子名称: | 58 kd capsid protein, ACETATE ION, MAGNESIUM ION, ... | | 著者 | Choi, J.M, Huston, A.M, Estes, M.K, Prasad, B.V.V. | | 登録日 | 2008-04-02 | | 公開日 | 2008-07-22 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2EHO
 
 | | Crystal structure of human GINS complex | | 分子名称: | DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, GINS complex subunit 3, ... | | 著者 | Choi, J.M, Lim, H.S, Kim, J.J, Song, O.K, Cho, Y. | | 登録日 | 2007-03-07 | | 公開日 | 2007-06-19 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure of the human GINS complex
Genes Dev., 21, 2007
|
|
1R6M
 
 | | Crystal Structure Of The tRNA Processing Enzyme Rnase pH From Pseudomonas Aeruginosa In Complex With Phosphate | | 分子名称: | PHOSPHATE ION, Ribonuclease PH | | 著者 | Choi, J.M, Park, E.Y, Kim, J.H, Chang, S.K, Cho, Y. | | 登録日 | 2003-10-15 | | 公開日 | 2004-02-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Probing the functional importance of the hexameric ring structure of RNase PH
J.BIOL.CHEM., 279, 2004
|
|
1R6L
 
 | | Crystal Structure Of The tRNA Processing Enzyme Rnase pH From Pseudomonas Aeruginosa | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Ribonuclease PH, SULFATE ION | | 著者 | Choi, J.M, Park, E.Y, Kim, J.H, Chang, S.K, Cho, Y. | | 登録日 | 2003-10-15 | | 公開日 | 2004-02-17 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Probing the functional importance of the hexameric ring structure of RNase PH
J.BIOL.CHEM., 279, 2004
|
|
3W0L
 
 | | The crystal structure of Xenopus Glucokinase and Glucokinase Regulatory Protein complex | | 分子名称: | FRUCTOSE -6-PHOSPHATE, Glucokinase, Glucokinase regulatory protein, ... | | 著者 | Choi, J.M, Seo, M.H, Kyeong, H.H, Kim, E, Kim, H.S. | | 登録日 | 2012-10-31 | | 公開日 | 2013-07-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.92 Å) | | 主引用文献 | Molecular basis for the role of glucokinase regulatory protein as the allosteric switch for glucokinase
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4R1P
 
 | |
5SV6
 
 | |
4R1Q
 
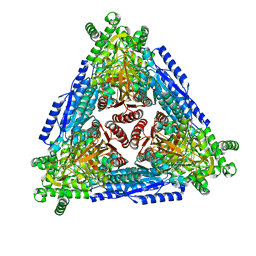 | |
4R1O
 
 | |
5GLC
 
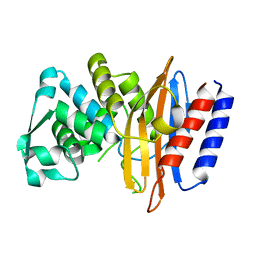 | | Crystal structure of the class A beta-lactamase PenL-tTR11 containing 20 residues insertion in omega-loop | | 分子名称: | Beta-lactamase | | 著者 | Choi, J.M, Yi, H, Kim, H.S, Lee, S.H. | | 登録日 | 2016-07-10 | | 公開日 | 2017-02-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.601 Å) | | 主引用文献 | High adaptability of the omega loop underlies the substrate-spectrum-extension evolution of a class A beta-lactamase, PenL
Sci Rep, 6, 2016
|
|
5GLD
 
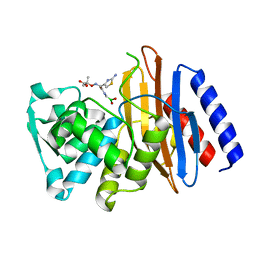 | | Crystal structure of the class A beta-lactamase PenL-tTR11 in complex with CBA | | 分子名称: | Beta-lactamase, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE | | 著者 | Choi, J.M, Yi, H, Kim, H.S, Lee, S.H. | | 登録日 | 2016-07-10 | | 公開日 | 2017-02-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | High adaptability of the omega loop underlies the substrate-spectrum-extension evolution of a class A beta-lactamase, PenL
Sci Rep, 6, 2016
|
|
5GLA
 
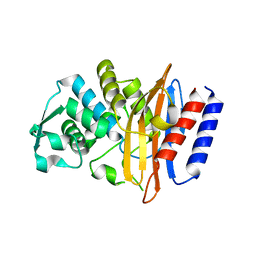 | | Crystal structure of the class A beta-lactamase PenL-tTR10 containing 10 residues insertion in omega-loop | | 分子名称: | Beta-lactamase | | 著者 | Choi, J.M, Yi, H, Kim, H.S, Lee, S.H. | | 登録日 | 2016-07-10 | | 公開日 | 2017-02-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | High adaptability of the omega loop underlies the substrate-spectrum-extension evolution of a class A beta-lactamase, PenL
Sci Rep, 6, 2016
|
|
5GL9
 
 | | Crystal structure of the class A beta-lactamase PenL | | 分子名称: | Beta-lactamase, GLYCEROL | | 著者 | Choi, J.M, Yi, H, Kim, H.S, Lee, S.H. | | 登録日 | 2016-07-10 | | 公開日 | 2017-02-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | High adaptability of the omega loop underlies the substrate-spectrum-extension evolution of a class A beta-lactamase, PenL
Sci Rep, 6, 2016
|
|
5GLB
 
 | | Crystal structure of the class A beta-lactamase PenL-tTR10 in complex with CBA | | 分子名称: | Beta-lactamase, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE | | 著者 | Choi, J.M, Yi, H, Kim, H.S, Lee, S.H. | | 登録日 | 2016-07-10 | | 公開日 | 2017-02-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | High adaptability of the omega loop underlies the substrate-spectrum-extension evolution of a class A beta-lactamase, PenL
Sci Rep, 6, 2016
|
|
4ZRM
 
 | |
4ZRN
 
 | |
5DBU
 
 | |
4OPH
 
 | | X-ray structure of full-length H6N6 NS1 | | 分子名称: | Nonstructural protein 1 | | 著者 | Carrillo, B, Choi, J.M, Bornholdt, Z.A, Sankaran, S, Rice, A.P, Prasad, B.V.V. | | 登録日 | 2014-02-05 | | 公開日 | 2014-02-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.158 Å) | | 主引用文献 | The Influenza A Virus Protein NS1 Displays Structural Polymorphism.
J.Virol., 88, 2014
|
|
2ZXX
 
 | | Crystal structure of Cdt1/geminin complex | | 分子名称: | DNA replication factor Cdt1, Geminin | | 著者 | Cho, Y, Lee, C, Hong, B.S, Choi, J.M. | | 登録日 | 2009-01-08 | | 公開日 | 2009-02-17 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis for inhibition of the replication licensing factor Cdt1 by geminin
Nature, 430, 2004
|
|
2ZIU
 
 | | Crystal structure of the Mus81-Eme1 complex | | 分子名称: | Crossover junction endonuclease EME1, Mus81 protein | | 著者 | Chang, J.H, Kim, J.J, Choi, J.M, Lee, J.H, Cho, Y. | | 登録日 | 2008-02-25 | | 公開日 | 2008-04-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of the Mus81-Eme1 complex
Genes Dev., 22, 2008
|
|
2ZIV
 
 | | Crystal structure of the Mus81-Eme1 complex | | 分子名称: | Crossover junction endonuclease EME1, Mus81 protein | | 著者 | Chang, J.H, Kim, J.J, Choi, J.M, Lee, J.H, Cho, Y. | | 登録日 | 2008-02-25 | | 公開日 | 2008-04-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of the Mus81-Eme1 complex
Genes Dev., 22, 2008
|
|
2ZIX
 
 | | Crystal structure of the Mus81-Eme1 complex | | 分子名称: | Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81 | | 著者 | Chang, J.H, Kim, J.J, Choi, J.M, Lee, J.H, Cho, Y. | | 登録日 | 2008-02-25 | | 公開日 | 2008-04-29 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Crystal structure of the Mus81-Eme1 complex
Genes Dev., 22, 2008
|
|
