1NMW
 
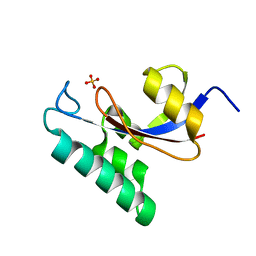 | | Solution structure of the PPIase domain of human Pin1 | | 分子名称: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, SULFATE ION | | 著者 | Bayer, E, Goettsch, S, Mueller, J.W, Griewel, B, Guiberman, E, Mayr, L, Bayer, P. | | 登録日 | 2003-01-12 | | 公開日 | 2003-07-15 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Analysis of the Mitotic Regulator hPin1 in Solution: INSIGHTS INTO DOMAIN ARCHITECTURE AND SUBSTRATE BINDING.
J.Biol.Chem., 278, 2003
|
|
1NMV
 
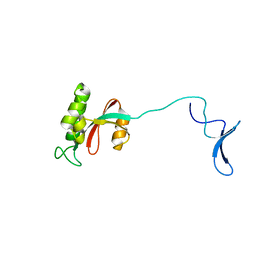 | | Solution structure of human Pin1 | | 分子名称: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Bayer, E, Goettsch, S, Mueller, J.W, Griewel, B, Guiberman, E, Mayr, L, Bayer, P. | | 登録日 | 2003-01-11 | | 公開日 | 2003-08-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Analysis of the Mitotic Regulator hPin1 in Solution: INSIGHTS INTO DOMAIN ARCHITECTURE AND SUBSTRATE BINDING.
J.Biol.Chem., 278, 2003
|
|
3BWZ
 
 | | Crystal structure of the type II cohesin module from the cellulosome of Acetivibrio cellulolyticus with an extended linker conformation | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Cellulosomal scaffoldin adaptor protein B, ... | | 著者 | Noach, I, Lamed, R, Shimon, L.J.W, Bayer, E, Frolow, F. | | 登録日 | 2008-01-10 | | 公開日 | 2009-01-13 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Intermodular linker flexibility revealed from crystal structures of adjacent cellulosomal cohesins of Acetivibrio cellulolyticus.
J.Mol.Biol., 391, 2009
|
|
1ZV9
 
 | | Crystal structure analysis of a type II cohesin domain from the cellulosome of Acetivibrio cellulolyticus- SeMet derivative | | 分子名称: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, ACETIC ACID, ... | | 著者 | Noach, I, Rosenheck, S, Lamed, R, Shimon, L, Bayer, E, Frolow, F. | | 登録日 | 2005-06-01 | | 公開日 | 2006-06-13 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | Intermodular linker flexibility revealed from crystal structures of adjacent cellulosomal cohesins of Acetivibrio cellulolyticus.
J.Mol.Biol., 391, 2009
|
|
5OGZ
 
 | |
4EYZ
 
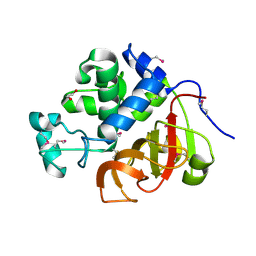 | | Crystal structure of an uncommon cellulosome-related protein module from Ruminococcus flavefaciens that resembles papain-like cysteine peptidases | | 分子名称: | 1,2-ETHANEDIOL, Cellulosome-related protein module from Ruminococcus flavefaciens that resembles papain-like cysteine peptidases | | 著者 | Frolow, F, Voronov-Goldman, M, Bayer, E, Lamed, R. | | 登録日 | 2012-05-02 | | 公開日 | 2013-03-20 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.383 Å) | | 主引用文献 | Crystal Structure of an Uncommon Cellulosome-Related Protein Module from Ruminococcus flavefaciens That Resembles Papain-Like Cysteine Peptidases.
Plos One, 8, 2013
|
|
4IU2
 
 | | Cohesin-dockerin -X domain complex from Ruminococcus flavefacience | | 分子名称: | CALCIUM ION, CHLORIDE ION, Cell-wall anchoring protein, ... | | 著者 | Salama-Alber, O, Bayer, E, Frolow, F. | | 登録日 | 2013-01-19 | | 公開日 | 2013-04-24 | | 最終更新日 | 2013-07-03 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Atypical Cohesin-Dockerin Complex Responsible for Cell Surface Attachment of Cellulosomal Components: BINDING FIDELITY, PROMISCUITY, AND STRUCTURAL BUTTRESSES.
J.Biol.Chem., 288, 2013
|
|
4IU3
 
 | | Cohesin-dockerin -X domain complex from Ruminococcus flavefacience | | 分子名称: | CALCIUM ION, Cell-wall anchoring protein, Cellulose-binding protein, ... | | 著者 | Salama-Alber, O, Bayer, E, Frolow, F. | | 登録日 | 2013-01-19 | | 公開日 | 2013-04-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Atypical Cohesin-Dockerin Complex Responsible for Cell Surface Attachment of Cellulosomal Components: BINDING FIDELITY, PROMISCUITY, AND STRUCTURAL BUTTRESSES.
J.Biol.Chem., 288, 2013
|
|
4N2O
 
 | | Structure of a novel autonomous cohesin protein from Ruminococcus flavefaciens | | 分子名称: | Autonomous cohesin, CHLORIDE ION | | 著者 | Frolow, F, Voronov-Goldman, M, Levy-Assaraf, M, Lamed, R, Bayer, E, Shimon, L. | | 登録日 | 2013-10-05 | | 公開日 | 2013-12-18 | | 最終更新日 | 2019-07-17 | | 実験手法 | X-RAY DIFFRACTION (2.442 Å) | | 主引用文献 | Structural characterization of a novel autonomous cohesin from Ruminococcus flavefaciens.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
2ZF9
 
 | |
7LSA
 
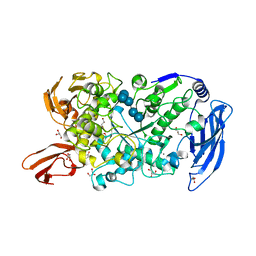 | | Ruminococcus bromii Amy12 with maltoheptaose | | 分子名称: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Koropatkin, N.M, Cockburn, D.W, Brown, H.A, Kibler, R.D. | | 登録日 | 2021-02-18 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Structure and substrate recognition by the Ruminococcus bromii amylosome pullulanases.
J.Struct.Biol., 213, 2021
|
|
7LST
 
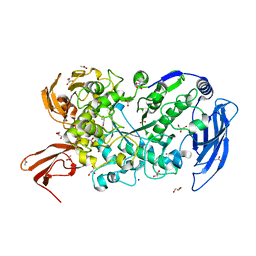 | | Ruminococcus bromii Amy12-D392A with 63-a-D-glucosyl-maltotriosyl-maltotriose | | 分子名称: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | 著者 | Koropatkin, N.M, Cockburn, D.W, Brown, H.A, Kibler, R.D. | | 登録日 | 2021-02-18 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure and substrate recognition by the Ruminococcus bromii amylosome pullulanases.
J.Struct.Biol., 213, 2021
|
|
7LSU
 
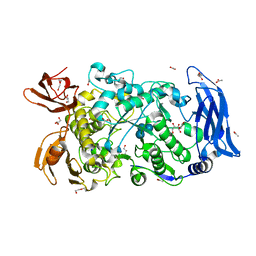 | | Ruminococcus bromii Amy12-D392A with 63-a-D-glucosyl-maltotriose | | 分子名称: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Koropatkin, N.M, Cockburn, D.W, Brown, H.A, Kibler, R.D. | | 登録日 | 2021-02-18 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure and substrate recognition by the Ruminococcus bromii amylosome pullulanases.
J.Struct.Biol., 213, 2021
|
|
7LSR
 
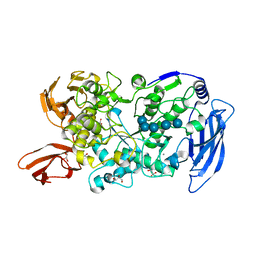 | | Ruminococcus bromii Amy12-D392A with maltoheptaose | | 分子名称: | CALCIUM ION, GLYCEROL, Pullulanase, ... | | 著者 | Koropatkin, N.M, Cockburn, D.W, Brown, H.A, Kibler, R.D. | | 登録日 | 2021-02-18 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | Structure and substrate recognition by the Ruminococcus bromii amylosome pullulanases.
J.Struct.Biol., 213, 2021
|
|
