4ZDU
 
 | |
5WUN
 
 | |
5WUM
 
 | |
5X8N
 
 | |
2LPU
 
 | | Solution structures of KmAtg10 | | 分子名称: | KmAtg10 | | 著者 | Yamaguchi, M, Noda, N.N, Yamamoto, H, Shima, T, Kumeta, H, Kobashigawa, Y, Akada, R, Ohsumi, Y, Inagaki, F. | | 登録日 | 2012-02-19 | | 公開日 | 2012-08-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural insights into atg10-mediated formation of the autophagy-essential atg12-atg5 conjugate
Structure, 20, 2012
|
|
3VX7
 
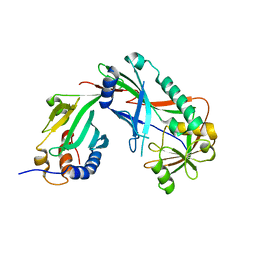 | | Crystal structure of Kluyveromyces marxianus Atg7NTD-Atg10 complex | | 分子名称: | E1, E2 | | 著者 | Yamaguchi, M, Matoba, K, Sawada, R, Fujioka, Y, Nakatogawa, H, Yamamoto, H, Kobashigawa, Y, Hoshida, H, Akada, R, Ohsumi, Y, Noda, N.N, Inagaki, F. | | 登録日 | 2012-09-11 | | 公開日 | 2012-11-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VX6
 
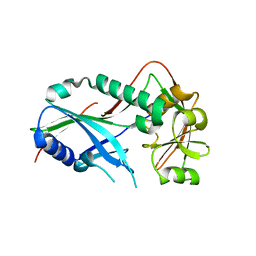 | | Crystal structure of Kluyveromyces marxianus Atg7NTD | | 分子名称: | E1 | | 著者 | Yamaguchi, M, Matoba, K, Sawada, R, Fujioka, Y, Nakatogawa, H, Yamamoto, H, Kobashigawa, Y, Hoshida, H, Akada, R, Ohsumi, Y, Noda, N.N, Inagaki, F. | | 登録日 | 2012-09-11 | | 公開日 | 2012-11-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VQI
 
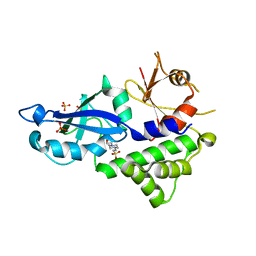 | | Crystal structure of Kluyveromyces marxianus Atg5 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Atg5, SULFATE ION | | 著者 | Yamaguchi, M, Noda, N.N, Yamamoto, H, Shima, T, Kumeta, H, Kobashigawa, Y, Akada, R, Ohsumi, Y, Inagaki, F. | | 登録日 | 2012-03-24 | | 公開日 | 2012-08-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural insights into atg10-mediated formation of the autophagy-essential atg12-atg5 conjugate
Structure, 20, 2012
|
|
1J2M
 
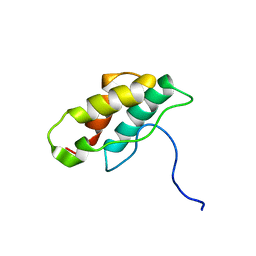 | | Solution structure of CPI-17(22-120) | | 分子名称: | 17-kDa PKC-potentiated inhibitory protein of PP1 | | 著者 | Ohki, S, Eto, M, Takada, R, Shimizu, M, Brautigan, D.L, Kainosho, M. | | 登録日 | 2003-01-07 | | 公開日 | 2003-06-17 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Distinctive Solution Conformation of Phosphatase Inhibitor CPI-17 Substituted with Aspartate at the Phosphorylation-site Threonine Residue
J.Mol.Biol., 326, 2003
|
|
1J2N
 
 | | Solution structure of CPI-17(22-120) T38D | | 分子名称: | 17-kDa PKC-potentiated inhibitory protein of PP1 | | 著者 | Ohki, S, Eto, M, Shimizu, M, Takada, R, Brautigan, D.L, Kainosho, M. | | 登録日 | 2003-01-07 | | 公開日 | 2003-06-17 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Distinctive Solution Conformation of Phosphatase Inhibitor CPI-17 Substituted with Aspartate at the Phosphorylation-site Threonine Residue
J.Mol.Biol., 326, 2003
|
|
4FS3
 
 | | Crystal structure of Staphylococcus aureus enoyl-ACP reductase in complex with NADP and AFN-1252 | | 分子名称: | Enoyl-[acyl-carrier-protein] reductase [NADPH] FabI, N-methyl-N-[(3-methyl-1-benzofuran-2-yl)methyl]-3-(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-3-yl)propanamide, [[(2R,3S,4R,5R)-5-(3-aminocarbonyl-4H-pyridin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methyl hydrogen phosphate | | 著者 | Kaplan, N, Yethon, J, Bardouniotis, E, Thalakada, R, Albert, M, Awrey, D.E, Romanov, V, Dorsey, M, Ramnauth, J, Clarke, T.E, Schmid, M.B, Berman, J, Pauls, H.W. | | 登録日 | 2012-06-26 | | 公開日 | 2012-09-19 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Mode of Action, In Vitro Activity, and In Vivo Efficacy of AFN-1252, a Selective Antistaphylococcal FabI Inhibitor.
Antimicrob.Agents Chemother., 56, 2012
|
|
3ACF
 
 | | Crystal Structure of Carbohydrate-Binding Module Family 28 from Clostridium josui Cel5A in a ligand-free form | | 分子名称: | Beta-1,4-endoglucanase, CALCIUM ION, SULFATE ION | | 著者 | Tsukimoto, K, Takada, R, Araki, Y, Suzuki, K, Karita, S, Wakagi, T, Shoun, H, Watanabe, T, Fushinobu, S. | | 登録日 | 2010-01-04 | | 公開日 | 2010-03-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Recognition of cellooligosaccharides by a family 28 carbohydrate-binding module.
Febs Lett., 584, 2010
|
|
3ACH
 
 | | Crystal Structure of Carbohydrate-Binding Module Family 28 from Clostridium josui Cel5A in complex with cellotetraose | | 分子名称: | Beta-1,4-endoglucanase, CALCIUM ION, PHOSPHATE ION, ... | | 著者 | Tsukimoto, K, Takada, R, Araki, Y, Suzuki, K, Karita, S, Wakagi, T, Shoun, H, Watanabe, T, Fushinobu, S. | | 登録日 | 2010-01-04 | | 公開日 | 2010-03-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Recognition of cellooligosaccharides by a family 28 carbohydrate-binding module.
Febs Lett., 584, 2010
|
|
3ACG
 
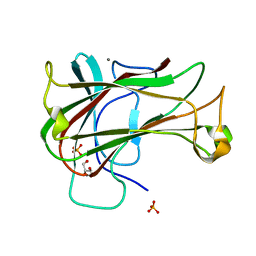 | | Crystal Structure of Carbohydrate-Binding Module Family 28 from Clostridium josui Cel5A in complex with cellobiose | | 分子名称: | Beta-1,4-endoglucanase, CALCIUM ION, GLYCEROL, ... | | 著者 | Tsukimoto, K, Takada, R, Araki, Y, Suzuki, K, Karita, S, Wakagi, T, Shoun, H, Watanabe, T, Fushinobu, S. | | 登録日 | 2010-01-04 | | 公開日 | 2010-03-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Recognition of cellooligosaccharides by a family 28 carbohydrate-binding module.
Febs Lett., 584, 2010
|
|
3ACI
 
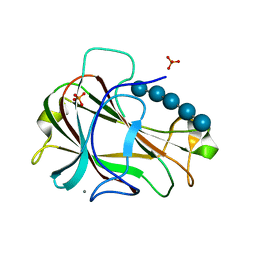 | | Crystal Structure of Carbohydrate-Binding Module Family 28 from Clostridium josui Cel5A in complex with cellopentaose | | 分子名称: | Beta-1,4-endoglucanase, CALCIUM ION, PHOSPHATE ION, ... | | 著者 | Tsukimoto, K, Takada, R, Araki, Y, Suzuki, K, Karita, S, Wakagi, T, Shoun, H, Watanabe, T, Fushinobu, S. | | 登録日 | 2010-01-04 | | 公開日 | 2010-03-31 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Recognition of cellooligosaccharides by a family 28 carbohydrate-binding module.
Febs Lett., 584, 2010
|
|
4P1W
 
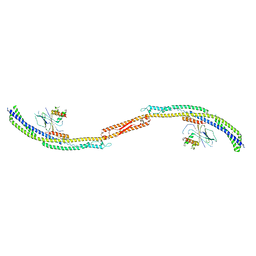 | |
4P1N
 
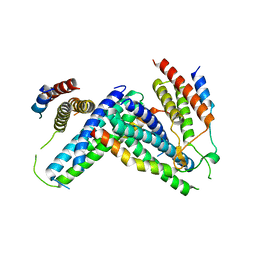 | |
3VX8
 
 | |
3VU4
 
 | |
