1GUH
 
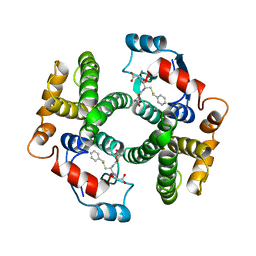 | | Structure determination and refinement of human alpha class glutathione transferase A1-1, and a comparison with the MU and PI class enzymes | | 分子名称: | GLUTATHIONE S-TRANSFERASE A1-1, S-BENZYL-GLUTATHIONE | | 著者 | Sinning, I, Kleywegt, G.J, Jones, T.A. | | 登録日 | 1993-02-24 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure determination and refinement of human alpha class glutathione transferase A1-1, and a comparison with the Mu and Pi class enzymes.
J.Mol.Biol., 232, 1993
|
|
1SUR
 
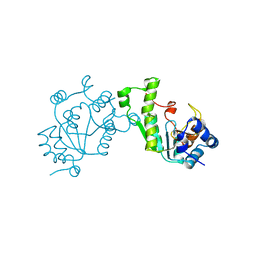 | | PHOSPHO-ADENYLYL-SULFATE REDUCTASE | | 分子名称: | PAPS REDUCTASE | | 著者 | Sinning, I, Savage, H. | | 登録日 | 1998-04-01 | | 公開日 | 1999-05-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of phosphoadenylyl sulphate (PAPS) reductase: a new family of adenine nucleotide alpha hydrolases.
Structure, 5, 1997
|
|
4CIU
 
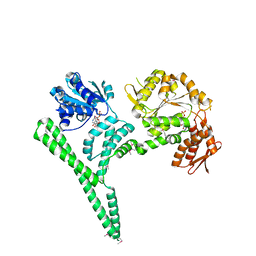 | | Crystal structure of E. coli ClpB | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CHAPERONE PROTEIN CLPB | | 著者 | Kopp, J, Sinning, I, Bukau, B, Kummer, E, Mogk, A. | | 登録日 | 2013-12-16 | | 公開日 | 2014-05-14 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Head-to-Tail Interactions of the Coiled-Coil Domains Regulate Clpb Cooperation with Hsp70 in Protein Disaggregation
Elife, 3, 2014
|
|
6TH0
 
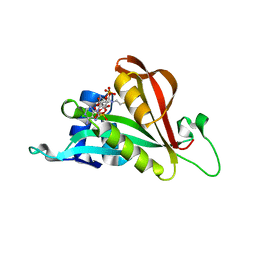 | | Crystal structure of Arabidopsis thaliana NAA60 in complex with acetyl-CoA | | 分子名称: | ACETYL COENZYME *A, Acyl-CoA N-acyltransferases (NAT) superfamily protein | | 著者 | Layer, D, Kopp, J, Lapouge, K, Sinning, I. | | 登録日 | 2019-11-18 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The Arabidopsis N alpha -acetyltransferase NAA60 locates to the plasma membrane and is vital for the high salt stress response.
New Phytol., 228, 2020
|
|
8CR1
 
 | | Homo sapiens Get1/Get2 heterotetramer in complex with a Get3 dimer | | 分子名称: | ATPase ASNA1, Guided entry of tail-anchored proteins factor CAMLG,Guided entry of tail-anchored proteins factor 1,GET2-GET1, ZINC ION | | 著者 | McDowell, M.A, Heimes, M, Wild, K, Sinning, I. | | 登録日 | 2023-03-07 | | 公開日 | 2023-11-29 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
4V7F
 
 | | Arx1 pre-60S particle. | | 分子名称: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | 著者 | Leidig, C, Thoms, M, Holdermann, I, Bradatsch, B, Berninghausen, O, Bange, G, Sinning, I, Hurt, E, Beckmann, R. | | 登録日 | 2013-12-10 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (8.7 Å) | | 主引用文献 | 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle.
Nat Commun, 5, 2014
|
|
1MHN
 
 | |
3SKQ
 
 | |
8CQZ
 
 | | Homo sapiens Get3 in complex with the Get1 cytoplasmic domain | | 分子名称: | ATPase ASNA1, Guided entry of tail-anchored proteins factor 1 | | 著者 | McDowell, M.A, Heimes, M, Wild, K, Saar, D, Sinning, I. | | 登録日 | 2023-03-07 | | 公開日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
4UE4
 
 | | Structural basis for targeting and elongation arrest of Bacillus signal recognition particle | | 分子名称: | 6S RNA, FTSQ SIGNAL SEQUENCE, SIGNAL RECOGNITION PARTICLE PROTEIN | | 著者 | Beckert, B, Kedrov, A, Sohmen, D, Kempf, G, Wild, K, Sinning, I, Stahlberg, H, Wilson, D.N, Beckmann, R. | | 登録日 | 2014-12-15 | | 公開日 | 2015-09-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7 Å) | | 主引用文献 | Translational Arrest by a Prokaryotic Signal Recognition Particle is Mediated by RNA Interactions.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UE5
 
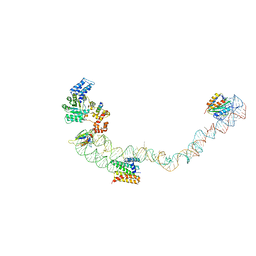 | | Structural basis for targeting and elongation arrest of Bacillus signal recognition particle | | 分子名称: | 7S RNA, SIGNAL RECOGNITION PARTICLE 54 KDA PROTEIN, SIGNAL RECOGNITION PARTICLE 9 KDA PROTEIN, ... | | 著者 | Beckert, B, Kedrov, A, Sohmen, D, Kempf, G, Wild, K, Sinning, I, Stahlberg, H, Wilson, D.N, Beckmann, R. | | 登録日 | 2014-12-15 | | 公開日 | 2015-09-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (9 Å) | | 主引用文献 | Translational Arrest by a Prokaryotic Signal Recognition Particle is Mediated by RNA Interactions.
Nat.Struct.Mol.Biol., 22, 2015
|
|
8QDQ
 
 | |
8QDR
 
 | | Vitis vinifera dimeric 13S-lipoxygenase LOXA in a detergent bound open conformation | | 分子名称: | 1,2-ETHANEDIOL, 3,6,12,15,18,21,24-HEPTAOXAHEXATRIACONTAN-1-OL, FE (III) ION, ... | | 著者 | Wild, K, Sinning, I. | | 登録日 | 2023-08-30 | | 公開日 | 2024-10-30 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Vitis vinifera Lipoxygenase LoxA is an Allosteric Dimer Activated by Lipidic Surfaces.
J.Mol.Biol., 436, 2024
|
|
8CR2
 
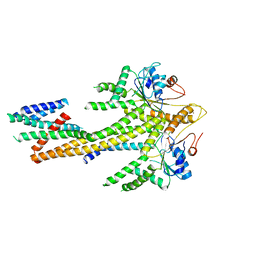 | | Homo sapiens Get1/Get2 heterotetramer (a3' deletion variant) in complex with a Get3 dimer | | 分子名称: | ATPase ASNA1, Guided entry of tail-anchored proteins factor CAMLG,Guided entry of tail-anchored proteins factor 1, ZINC ION | | 著者 | McDowell, M.A, Heimes, M, Wild, K, Sinning, I. | | 登録日 | 2023-03-07 | | 公開日 | 2023-11-29 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
4NWB
 
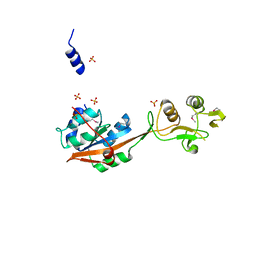 | | Crystal structure of Mrt4 | | 分子名称: | SULFATE ION, mRNA turnover protein 4 | | 著者 | Holdermann, I, Sinning, I. | | 登録日 | 2013-12-06 | | 公開日 | 2014-03-26 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle.
Nat Commun, 5, 2014
|
|
6EMG
 
 | |
4IPA
 
 | | Structure of a thermophilic Arx1 | | 分子名称: | Putative curved DNA-binding protein, SULFATE ION | | 著者 | Bange, G, Sinning, I. | | 登録日 | 2013-01-09 | | 公開日 | 2013-01-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Consistent mutational paths predict eukaryotic thermostability.
BMC Evol Biol, 13, 2013
|
|
5TKY
 
 | | Crystal structure of the co-translational Hsp70 chaperone Ssb in the ATP-bound, open conformation | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Putative uncharacterized protein | | 著者 | Gumiero, A, Gese, G.V, Weyer, F.A, Lapouge, K, Sinning, I. | | 登録日 | 2016-10-10 | | 公開日 | 2016-11-16 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Interaction of the cotranslational Hsp70 Ssb with ribosomal proteins and rRNA depends on its lid domain.
Nat Commun, 7, 2016
|
|
1Y10
 
 | | Mycobacterial adenylyl cyclase Rv1264, holoenzyme, inhibited state | | 分子名称: | CALCIUM ION, Hypothetical protein Rv1264/MT1302, PENTAETHYLENE GLYCOL | | 著者 | Tews, I, Findeisen, F, Sinning, I, Schultz, A, Schultz, J.E, Linder, J.U. | | 登録日 | 2004-11-16 | | 公開日 | 2005-05-24 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The structure of a pH-sensing mycobacterial adenylyl cyclase holoenzyme
Science, 308, 2005
|
|
1Y11
 
 | | Mycobacterial adenylyl cyclase Rv1264, holoenzyme, active state | | 分子名称: | GLYCEROL, Hypothetical protein Rv1264/MT1302, PENTAETHYLENE GLYCOL, ... | | 著者 | Tews, I, Findeisen, F, Sinning, I, Schultz, A, Schultz, J.E, Linder, J.U. | | 登録日 | 2004-11-16 | | 公開日 | 2005-05-24 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | The structure of a pH-sensing mycobacterial adenylyl cyclase holoenzyme
Science, 308, 2005
|
|
8QL1
 
 | | Crystal structure of the human MDN1-MIDAS/NLE1-UBL complex | | 分子名称: | MAGNESIUM ION, NDE1, Notchless protein homolog 1 | | 著者 | Wild, K, Fiorentino, F, Hurt, E, Sinning, I. | | 登録日 | 2023-09-19 | | 公開日 | 2025-04-09 | | 最終更新日 | 2025-04-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Highly conserved ribosome biogenesis pathways between human and yeast revealed by the MDN1-NLE1 interaction and NLE1 containing pre-60S subunits.
Nucleic Acids Res., 53, 2025
|
|
1FTS
 
 | |
3SJC
 
 | |
3SJA
 
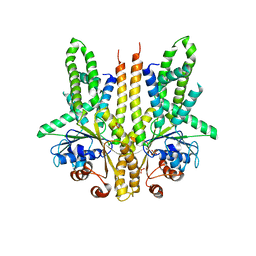 | | Crystal structure of S. cerevisiae Get3 in the open state in complex with Get1 cytosolic domain | | 分子名称: | ATPase GET3, Golgi to ER traffic protein 1, PHOSPHATE ION, ... | | 著者 | Reitz, S, Wild, K, Sinning, I. | | 登録日 | 2011-06-21 | | 公開日 | 2011-07-06 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex.
Science, 333, 2011
|
|
3SJD
 
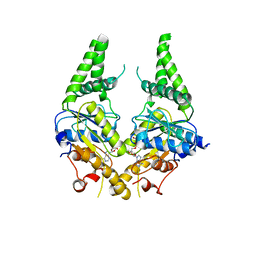 | | Crystal structure of S. cerevisiae Get3 with bound ADP-Mg2+ in complex with Get2 cytosolic domain | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATPase GET3, Golgi to ER traffic protein 2, ... | | 著者 | Reitz, S, Wild, K, Sinning, I. | | 登録日 | 2011-06-21 | | 公開日 | 2011-07-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (4.6 Å) | | 主引用文献 | Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex.
Science, 333, 2011
|
|
