6MSH
 
 | |
6VYO
 
 | | Crystal structure of RNA binding domain of nucleocapsid phosphoprotein from SARS coronavirus 2 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | 著者 | Chang, C, Michalska, K, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Kim, Y, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-02-27 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
8K0R
 
 | |
8K15
 
 | |
8RJX
 
 | |
7XMZ
 
 | |
7XMX
 
 | |
6MSE
 
 | |
6MSB
 
 | |
8KHD
 
 | | The interface structure of Omicron RBD binding to 5817 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 5817, Light chain of 5817, ... | | 著者 | Cao, L, Wang, X. | | 登録日 | 2023-08-21 | | 公開日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Identification of a broad sarbecovirus neutralizing antibody targeting a conserved epitope on the receptor-binding domain.
Cell Rep, 43, 2024
|
|
8KHC
 
 | | SARS-CoV-2 Omicron spike in complex with 5817 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 5817 Fab, ... | | 著者 | Cao, L, Wang, X. | | 登録日 | 2023-08-21 | | 公開日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Identification of a broad sarbecovirus neutralizing antibody targeting a conserved epitope on the receptor-binding domain.
Cell Rep, 43, 2024
|
|
8K0S
 
 | |
8K0Q
 
 | |
8K0P
 
 | |
6VQ2
 
 | | HLA-B*27:05 presenting an HIV-1 14mer peptide | | 分子名称: | 14-mer peptide, Beta-2-microglobulin, GLYCEROL, ... | | 著者 | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | 登録日 | 2020-02-04 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
6VQD
 
 | | HLA-B*27:05 presenting an HIV-1 8mer peptide | | 分子名称: | 8-mer peptide, Beta-2-microglobulin, GLYCEROL, ... | | 著者 | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | 登録日 | 2020-02-05 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
6VQE
 
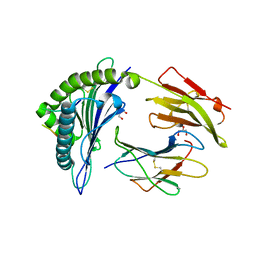 | | HLA-B*27:05 presenting an HIV-1 13mer peptide | | 分子名称: | 13-mer peptide, Beta-2-microglobulin, GLYCEROL, ... | | 著者 | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | 登録日 | 2020-02-05 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
6VQY
 
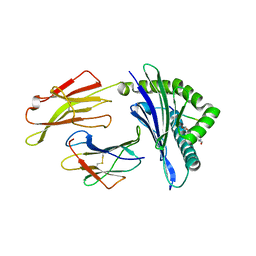 | | HLA-B*27:05 presenting an HIV-1 7mer peptide | | 分子名称: | 7-mer peptide, ARGININE, Beta-2-microglobulin, ... | | 著者 | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | 登録日 | 2020-02-06 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
6VQZ
 
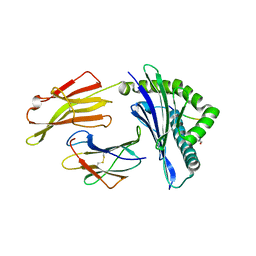 | | HLA-B*27:05 presenting an HIV-1 6mer peptide | | 分子名称: | 6-mer peptide, ARGININE, Beta-2-microglobulin, ... | | 著者 | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | 登録日 | 2020-02-06 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
6VPZ
 
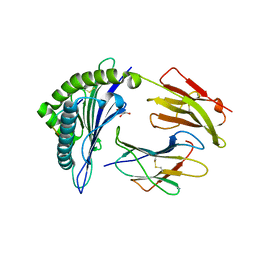 | | HLA-B*27:05 presenting an HIV-1 11mer peptide | | 分子名称: | 11-mer peptide, Beta-2-microglobulin, GLYCEROL, ... | | 著者 | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | 登録日 | 2020-02-04 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
8JEY
 
 | |
4X1I
 
 | | Discovery of cytotoxic Dolastatin 10 analogs with N-terminal modifications | | 分子名称: | 2-methyl-L-alanyl-N-[(3R,4S,5S)-3-methoxy-1-{(2S)-2-[(1R,2R)-1-methoxy-2-methyl-3-oxo-3-{[(1S)-2-phenyl-1-(1,3-thiazol-2-yl)ethyl]amino}propyl]pyrrolidin-1-yl}-5-methyl-1-oxoheptan-4-yl]-N-methyl-L-valinamide, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Parris, K.D. | | 登録日 | 2014-11-24 | | 公開日 | 2015-03-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.11 Å) | | 主引用文献 | Discovery of cytotoxic dolastatin 10 analogues with N-terminal modifications.
J.Med.Chem., 57, 2014
|
|
6LN4
 
 | | Estrogen-related receptor beta(ERR2) in complex with PGC1a-2a | | 分子名称: | 10-mer from Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, Steroid hormone receptor ERR2 | | 著者 | Yao, B.Q, Li, Y. | | 登録日 | 2019-12-28 | | 公開日 | 2020-10-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Structural Insights into the Specificity of Ligand Binding and Coactivator Assembly by Estrogen-Related Receptor beta.
J.Mol.Biol., 432, 2020
|
|
4X1Y
 
 | |
8J1H
 
 | | Agonist1 and Ruthenium Red bound state of mTRPV4 | | 分子名称: | 8-fluoranyl-3-[4-(4-fluoranylphenoxy)phenyl]-2-(4-methylpiperazin-1-yl)quinazolin-4-one, Transient receptor potential cation channel subfamily V member 4 | | 著者 | Zhen, W.X, Yang, F. | | 登録日 | 2023-04-12 | | 公開日 | 2023-08-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.88 Å) | | 主引用文献 | Structural basis of ligand activation and inhibition in a mammalian TRPV4 ion channel.
Cell Discov, 9, 2023
|
|
