2D23
 
 | | Crystal structure of EP complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | 分子名称: | AZIDE ION, ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, ... | | 著者 | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | 登録日 | 2005-09-02 | | 公開日 | 2006-10-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
2D1Z
 
 | | Crystal structure of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | 分子名称: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, SULFATE ION | | 著者 | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | 登録日 | 2005-09-02 | | 公開日 | 2006-10-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
2D20
 
 | | Crystal structure of michaelis complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | 分子名称: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, P-NITROPHENOL, ... | | 著者 | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | 登録日 | 2005-09-02 | | 公開日 | 2006-10-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
2D24
 
 | | Crystal structure of ES complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | 分子名称: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, SULFATE ION, ... | | 著者 | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | 登録日 | 2005-09-02 | | 公開日 | 2006-10-10 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
6JUD
 
 | | Radiation damage in Aspergillus oryzae pro-tyrosinase oxygen-bound C92A/H103F mutant | | 分子名称: | COPPER (II) ION, PEROXIDE ION, Tyrosinase | | 著者 | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | 登録日 | 2019-04-13 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6JU6
 
 | | Aspergillus oryzae active-tyrosinase copper-depleted C92A mutant | | 分子名称: | NITRATE ION, Tyrosinase | | 著者 | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | 登録日 | 2019-04-13 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6JUC
 
 | | Aspergillus oryzae pro-tyrosinase oxygen-bound C92A/H103F mutant | | 分子名称: | COPPER (II) ION, PEROXIDE ION, Tyrosinase | | 著者 | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | 登録日 | 2019-04-13 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6JU5
 
 | | Aspergillus oryzae pro-tyrosinase C92A/F513Y mutant | | 分子名称: | COPPER (II) ION, Tyrosinase | | 著者 | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | 登録日 | 2019-04-13 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6JU9
 
 | | Aspergillus oryzae active-tyrosinase copper-bound C92A mutant complexed with L-tyrosine | | 分子名称: | 3,4-DIHYDROXYPHENYLALANINE, COPPER (II) ION, NITRATE ION, ... | | 著者 | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | 登録日 | 2019-04-13 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6JU7
 
 | | Aspergillus oryzae active-tyrosinase copper-depleted C92A mutant complexed with L-tyrosine | | 分子名称: | NITRATE ION, TYROSINE, Tyrosinase | | 著者 | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | 登録日 | 2019-04-13 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
2DIE
 
 | | Alkaline alpha-amylase AmyK from Bacillus sp. KSM-1378 | | 分子名称: | CALCIUM ION, SODIUM ION, amylase | | 著者 | Shirai, T, Igarashi, K, Ozawa, T, Hagihara, H, Kobayashi, T, Ozaki, K, Ito, S. | | 登録日 | 2006-03-29 | | 公開日 | 2007-02-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Ancestral sequence evolutionary trace and crystal structure analyses of alkaline alpha-amylase from Bacillus sp. KSM-1378 to clarify the alkaline adaptation process of proteins
Proteins, 66, 2007
|
|
6JUA
 
 | | Aspergillus oryzae pro-tyrosinase oxygen-bound C92A mutant | | 分子名称: | COPPER (II) ION, PEROXIDE ION, Tyrosinase | | 著者 | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | 登録日 | 2019-04-13 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6JU4
 
 | | Aspergillus oryzae pro-tyrosinase F513Y mutant | | 分子名称: | COPPER (II) ION, Tyrosinase | | 著者 | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | 登録日 | 2019-04-13 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6JU8
 
 | | Aspergillus oryzae active-tyrosinase copper-bound C92A mutant | | 分子名称: | COPPER (II) ION, NITRATE ION, Tyrosinase | | 著者 | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | 登録日 | 2019-04-13 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.27 Å) | | 主引用文献 | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6JUB
 
 | | Radiation damage in Aspergillus oryzae pro-tyrosinase oxygen-bound C92A mutant | | 分子名称: | COPPER (II) ION, PEROXIDE ION, Tyrosinase | | 著者 | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | 登録日 | 2019-04-13 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
1EE6
 
 | | CRYSTAL STRUCTURE OF PECTATE LYASE FROM BACILLUS SP. STRAIN KSM-P15. | | 分子名称: | CALCIUM ION, PECTATE LYASE | | 著者 | Akita, M, Suzuki, A, Kobayashi, T, Ito, S, Yamane, T. | | 登録日 | 2000-01-31 | | 公開日 | 2001-01-31 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The first structure of pectate lyase belonging to polysaccharide lyase family 3.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1G0C
 
 | | ALKALINE CELLULASE K CATALYTIC DOMAIN-CELLOBIOSE COMPLEX | | 分子名称: | ACETIC ACID, CADMIUM ION, ENDOGLUCANASE, ... | | 著者 | Shirai, T, Ishida, H, Noda, J, Yamane, T, Ozaki, K, Hakamada, Y, Ito, S. | | 登録日 | 2000-10-05 | | 公開日 | 2001-08-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of alkaline cellulase K: insight into the alkaline adaptation of an industrial enzyme.
J.Mol.Biol., 310, 2001
|
|
1G01
 
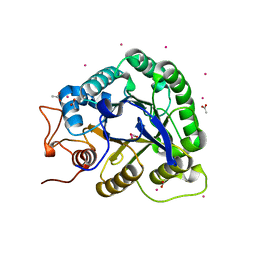 | | ALKALINE CELLULASE K CATALYTIC DOMAIN | | 分子名称: | ACETIC ACID, CADMIUM ION, ENDOGLUCANASE | | 著者 | Shirai, T, Ishida, H, Noda, J, Yamane, T, Ozaki, K, Hakamada, Y, Ito, S. | | 登録日 | 2000-10-05 | | 公開日 | 2001-08-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of alkaline cellulase K: insight into the alkaline adaptation of an industrial enzyme.
J.Mol.Biol., 310, 2001
|
|
1CTO
 
 | | NMR STRUCTURE OF THE C-TERMINAL DOMAIN OF THE LIGAND-BINDING REGION OF MURINE GRANULOCYTE COLONY-STIMULATING FACTOR RECEPTOR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | GRANULOCYTE COLONY-STIMULATING FACTOR RECEPTOR | | 著者 | Yamasaki, K, Naito, S, Anaguchi, H, Ohkubo, T, Ota, Y. | | 登録日 | 1996-09-25 | | 公開日 | 1997-10-22 | | 最終更新日 | 2018-03-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of an extracellular domain containing the WSxWS motif of the granulocyte colony-stimulating factor receptor and its interaction with ligand.
Nat.Struct.Biol., 4, 1997
|
|
3WAT
 
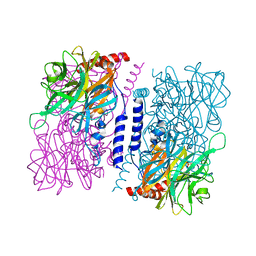 | | Crystal structure of 4-O-beta-D-mannosyl-D-glucose phosphorylase MGP complexed with Man+Glc | | 分子名称: | 4-O-beta-D-mannosyl-D-glucose phosphorylase, PHOSPHATE ION, beta-D-glucopyranose, ... | | 著者 | Nakae, S, Ito, S, Higa, M, Senoura, T, Wasaki, J, Hijikata, A, Shionyu, M, Ito, S, Shirai, T. | | 登録日 | 2013-05-08 | | 公開日 | 2013-09-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure of Novel Enzyme in Mannan Biodegradation Process 4-O-beta-d-Mannosyl-d-Glucose Phosphorylase MGP
J.Mol.Biol., 425, 2013
|
|
7X7K
 
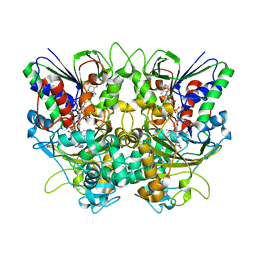 | | Ancestral L-Lys oxidase (AncLLysO-2) L-Arg binding form | | 分子名称: | ARGININE, FAD dependent enzyme, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Motoyama, T, Ishida, C, Hasebe, F, Ito, S, Nakano, S. | | 登録日 | 2022-03-09 | | 公開日 | 2023-01-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Reaction Mechanism of Ancestral l-Lys alpha-Oxidase from Caulobacter Species Studied by Biochemical, Structural, and Computational Analysis
Acs Omega, 7, 2022
|
|
7X7J
 
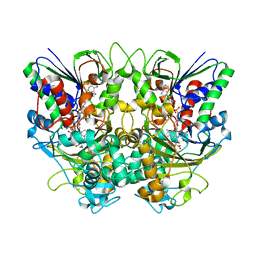 | | Ancestral L-Lys oxidase (AncLLysO-2) L-Lys binding form | | 分子名称: | FAD dependent enzyme, FLAVIN-ADENINE DINUCLEOTIDE, LYSINE | | 著者 | Motoyama, T, Ishida, C, Hasebe, F, Ito, S, Nakano, S. | | 登録日 | 2022-03-09 | | 公開日 | 2023-01-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Reaction Mechanism of Ancestral l-Lys alpha-Oxidase from Caulobacter Species Studied by Biochemical, Structural, and Computational Analysis
Acs Omega, 7, 2022
|
|
7X7I
 
 | | Ancestral L-Lys oxidase (AncLLysO-2) ligand free form | | 分子名称: | FAD dependent enzyme, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Motoyama, T, Ishida, C, Hasebe, F, Ito, S, Nakano, S. | | 登録日 | 2022-03-09 | | 公開日 | 2023-01-18 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Reaction Mechanism of Ancestral l-Lys alpha-Oxidase from Caulobacter Species Studied by Biochemical, Structural, and Computational Analysis
Acs Omega, 7, 2022
|
|
3WAU
 
 | | Crystal structure of 4-O-beta-D-mannosyl-D-glucose phosphorylase MGP complexed with M1P | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-O-phosphono-alpha-D-mannopyranose, 4-O-beta-D-mannosyl-D-glucose phosphorylase, ... | | 著者 | Nakae, S, Ito, S, Higa, M, Senoura, T, Wasaki, J, Hijikata, A, Shionyu, M, Ito, S, Shirai, T. | | 登録日 | 2013-05-08 | | 公開日 | 2013-09-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure of Novel Enzyme in Mannan Biodegradation Process 4-O-beta-d-Mannosyl-d-Glucose Phosphorylase MGP
J.Mol.Biol., 425, 2013
|
|
3WAS
 
 | | Crystal structure of 4-O-beta-D-mannosyl-D-glucose phosphorylase MGP complexed with Man-Glc+PO4 | | 分子名称: | 4-O-beta-D-mannosyl-D-glucose phosphorylase, PHOSPHATE ION, beta-D-mannopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Nakae, S, Ito, S, Higa, M, Senoura, T, Wasaki, J, Hijikata, A, Shionyu, M, Ito, S, Shirai, T. | | 登録日 | 2013-05-08 | | 公開日 | 2013-09-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure of Novel Enzyme in Mannan Biodegradation Process 4-O-beta-d-Mannosyl-d-Glucose Phosphorylase MGP
J.Mol.Biol., 425, 2013
|
|
