6JQ2
 
 | | Crystal Structure of H2-Kb in complex with a DPAGT1 self-peptide | | 分子名称: | Beta-2-microglobulin, DPATG1 antigen SIIVFNLV, H-2 class I histocompatibility antigen, ... | | 著者 | Bai, P, Yin, L. | | 登録日 | 2019-03-28 | | 公開日 | 2020-04-01 | | 最終更新日 | 2021-02-17 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Immune-based mutation classification enables neoantigen prioritization and immune feature discovery in cancer immunotherapy.
Oncoimmunology, 10, 2021
|
|
6JQ3
 
 | | Crystal Structure of H2-Kb in complex with a DPAGT1 mutant peptide | | 分子名称: | Beta-2-microglobulin, DPAGT1 mutant antigen SIIVFNLL, H-2 class I histocompatibility antigen, ... | | 著者 | Bai, P, Zhou, Q, Wei, P, Yin, L. | | 登録日 | 2019-03-28 | | 公開日 | 2020-05-06 | | 最終更新日 | 2021-02-17 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Immune-based mutation classification enables neoantigen prioritization and immune feature discovery in cancer immunotherapy.
Oncoimmunology, 10, 2021
|
|
6JTN
 
 | | Crystal structure of HLA-C08 in complex with a tumor mut10m peptide | | 分子名称: | 10-mer peptide, Beta-2-microglobulin, HLA class I antigen, ... | | 著者 | Bai, P, Zhou, Q, Wei, P, Yin, L. | | 登録日 | 2019-04-11 | | 公開日 | 2020-04-15 | | 最終更新日 | 2021-02-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Immune-based mutation classification enables neoantigen prioritization and immune feature discovery in cancer immunotherapy.
Oncoimmunology, 10, 2021
|
|
7XML
 
 | |
3QK5
 
 | | Crystal structure of fatty acid amide hydrolase with small molecule inhibitor | | 分子名称: | (3-{(3R)-1-[4-(1-benzothiophen-2-yl)pyrimidin-2-yl]piperidin-3-yl}-2-methyl-1H-pyrrolo[2,3-b]pyridin-1-yl)acetonitrile, 1,2-ETHANEDIOL, Fatty-acid amide hydrolase 1, ... | | 著者 | Min, X, Walker, N.P.C, Wang, Z. | | 登録日 | 2011-01-31 | | 公開日 | 2011-04-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Identification of potent, noncovalent fatty acid amide hydrolase (FAAH) inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6KBU
 
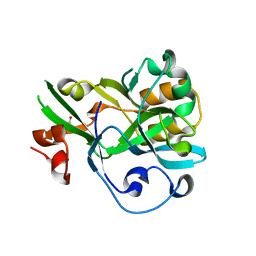 | | Crystal structure of yedK | | 分子名称: | GLYCEROL, SOS response-associated protein | | 著者 | Wang, N, Bao, H, Huang, H. | | 登録日 | 2019-06-26 | | 公開日 | 2019-07-10 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Molecular basis of abasic site sensing in single-stranded DNA by the SRAP domain of E. coli yedK.
Nucleic Acids Res., 47, 2019
|
|
4KTE
 
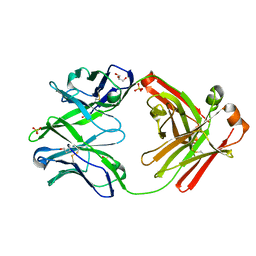 | | Fab fragment of HIV vaccine-elicited CD4bs-directed antibody, GE148, from non-human primate | | 分子名称: | GE148 Heavy Chain Fab, GE148 Light Chain Fab, GLYCEROL, ... | | 著者 | Poulsen, C, Tran, K, Stanfield, R, Wyatt, R.T. | | 登録日 | 2013-05-20 | | 公開日 | 2014-02-05 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Vaccine-elicited primate antibodies use a distinct approach to the HIV-1 primary receptor binding site informing vaccine redesign.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3QKV
 
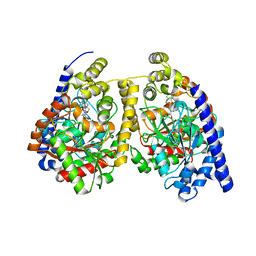 | | Crystal structure of fatty acid amide hydrolase with small molecule compound | | 分子名称: | (6-bromo-1'H,4H-spiro[1,3-benzodioxine-2,4'-piperidin]-1'-yl)methanol, Fatty-acid amide hydrolase 1 | | 著者 | Min, X, Walker, N.P.C, Wang, Z. | | 登録日 | 2011-02-01 | | 公開日 | 2011-04-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Discovery and molecular basis of potent noncovalent inhibitors of fatty acid amide hydrolase (FAAH).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3QJ9
 
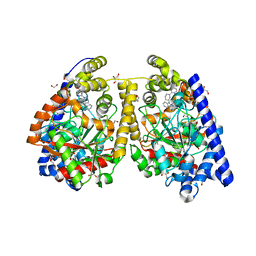 | | Crystal structure of fatty acid amide hydrolase with small molecule inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 1-{(3S)-1-[4-(1-benzofuran-2-yl)pyrimidin-2-yl]piperidin-3-yl}-3-ethyl-1,3-dihydro-2H-benzimidazol-2-one, Fatty-acid amide hydrolase 1, ... | | 著者 | Min, X, Walker, N.P.C, Wang, Z. | | 登録日 | 2011-01-28 | | 公開日 | 2011-04-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Discovery and molecular basis of potent noncovalent inhibitors of fatty acid amide hydrolase (FAAH).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6LQI
 
 | | Cryo-EM structure of the mouse Piezo1 isoform Piezo1.1 | | 分子名称: | Piezo-type mechanosensitive ion channel component 1 | | 著者 | Geng, J, Liu, W, Zhou, H, Zhang, T, Wang, L, Zhang, M, Shen, B, Li, X, Xiao, B. | | 登録日 | 2020-01-13 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | A Plug-and-Latch Mechanism for Gating the Mechanosensitive Piezo Channel.
Neuron, 106, 2020
|
|
5ZNY
 
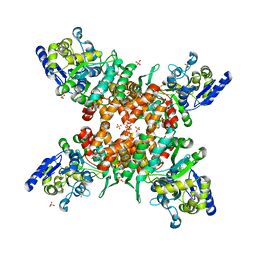 | | Structure of mDR3_DD-C363G with MBP tag | | 分子名称: | Maltose-binding periplasmic protein,Tumor necrosis factor receptor superfamily, member 25, SULFATE ION | | 著者 | Yin, X, Jin, T. | | 登録日 | 2018-04-11 | | 公開日 | 2019-04-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.74 Å) | | 主引用文献 | Crystal structure and activation mechanism of DR3 death domain.
Febs J., 286, 2019
|
|
5ZNZ
 
 | | Structure of mDR3 DD with MBP tag mutant-I387V | | 分子名称: | Maltose-binding periplasmic protein,Tumor necrosis factor receptor superfamily, member 25, SULFATE ION | | 著者 | Jin, T, Yin, X. | | 登録日 | 2018-04-12 | | 公開日 | 2019-04-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Crystal structure and activation mechanism of DR3 death domain.
Febs J., 286, 2019
|
|
2F54
 
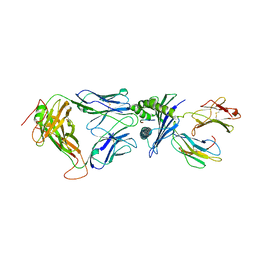 | | Directed evolution of human T cell receptor CDR2 residues by phage display dramatically enhances affinity for cognate peptide-MHC without increasing apparent cross-reactivity | | 分子名称: | Beta-2-microglobulin, Cancer/testis antigen 1B, HLA class I histocompatibility antigen, ... | | 著者 | Rizkallah, P.J, Jakobsen, B.K, Dunn, S.M, Sami, M. | | 登録日 | 2005-11-25 | | 公開日 | 2006-04-25 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Directed evolution of human T cell receptor CDR2 residues by phage display dramatically enhances affinity for cognate peptide-MHC without increasing apparent cross-reactivity.
Protein Sci., 15, 2006
|
|
6KIX
 
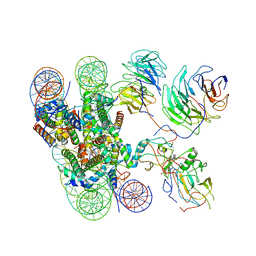 | | Cryo-EM structure of human MLL1-NCP complex, binding mode1 | | 分子名称: | DNA (145-MER), GLUTAMINE, Histone H2A, ... | | 著者 | Huang, J, Xue, H, Yao, T. | | 登録日 | 2019-07-20 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KIV
 
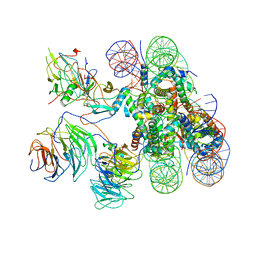 | | Cryo-EM structure of human MLL1-ubNCP complex (4.0 angstrom) | | 分子名称: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | 著者 | Huang, J, Xue, H, Yao, T. | | 登録日 | 2019-07-20 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KIU
 
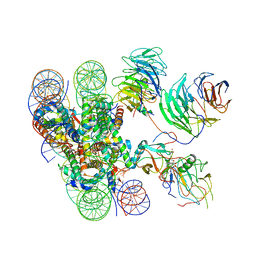 | | Cryo-EM structure of human MLL1-ubNCP complex (3.2 angstrom) | | 分子名称: | DNA (145-MER), GLUTAMINE, Histone H2A, ... | | 著者 | Huang, J, Xue, H, Yao, T. | | 登録日 | 2019-07-20 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KIJ
 
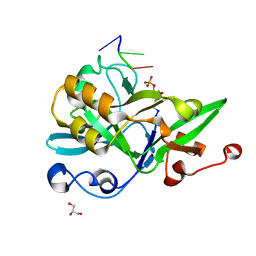 | | Crystal structure of yedK with ssDNA containing an abasic site | | 分子名称: | DNA (5'-D(*GP*AP*TP*TP*CP*GP*TP*CP*G)-3'), GLYCEROL, PENTANE-3,4-DIOL-5-PHOSPHATE, ... | | 著者 | Wang, N, Bao, H, Huang, H. | | 登録日 | 2019-07-18 | | 公開日 | 2019-08-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Molecular basis of abasic site sensing in single-stranded DNA by the SRAP domain of E. coli yedK.
Nucleic Acids Res., 47, 2019
|
|
6KIZ
 
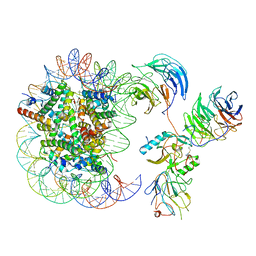 | | Cryo-EM structure of human MLL1-NCP complex, binding mode2 | | 分子名称: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | 著者 | Huang, J, Xue, H, Yao, T. | | 登録日 | 2019-07-20 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
2FLO
 
 | |
6KBZ
 
 | |
6KIW
 
 | | Cryo-EM structure of human MLL3-ubNCP complex (4.0 angstrom) | | 分子名称: | DNA (144-MER), DNA (145-MER), Histone H2A, ... | | 著者 | Huang, J, Xue, H, Yao, T. | | 登録日 | 2019-07-20 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
4NW3
 
 | | Crystal structure of MLL CXXC domain in complex with a CpG DNA | | 分子名称: | 5'-D(*GP*CP*CP*AP*TP*CP*GP*AP*TP*GP*GP*C)-3', Histone-lysine N-methyltransferase 2A, ZINC ION | | 著者 | Bian, C, Tempel, W, Chao, X, Walker, J.R, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2013-12-05 | | 公開日 | 2014-04-23 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.82 Å) | | 主引用文献 | DNA Sequence Recognition of Human CXXC Domains and Their Structural Determinants.
Structure, 26, 2018
|
|
4O64
 
 | | Zinc fingers of KDM2B | | 分子名称: | Lysine-specific demethylase 2B, UNKNOWN ATOM OR ION, ZINC ION | | 著者 | Liu, K, Xu, C, Tempel, W, Walker, J.R, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2013-12-20 | | 公開日 | 2014-04-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | DNA Sequence Recognition of Human CXXC Domains and Their Structural Determinants.
Structure, 26, 2018
|
|
3RQ4
 
 | | Crystal structure of suppressor of variegation 4-20 homolog 2 | | 分子名称: | 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SUV420H2, S-ADENOSYLMETHIONINE, ... | | 著者 | Dong, A, Zeng, H, Tempel, W, Loppnau, P, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | 登録日 | 2011-04-27 | | 公開日 | 2011-06-01 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structures of the human histone H4K20 methyltransferases SUV420H1 and SUV420H2.
Febs Lett., 587, 2013
|
|
3S8P
 
 | | Crystal Structure of the SET Domain of Human Histone-Lysine N-Methyltransferase SUV420H1 In Complex With S-Adenosyl-L-Methionine | | 分子名称: | Histone-lysine N-methyltransferase SUV420H1, S-ADENOSYLMETHIONINE, ZINC ION | | 著者 | Lam, R, Zeng, H, Loppnau, P, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | 登録日 | 2011-05-30 | | 公開日 | 2011-07-06 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structures of the human histone H4K20 methyltransferases SUV420H1 and SUV420H2.
Febs Lett., 587, 2013
|
|
