8XOI
 
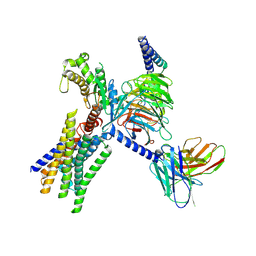 | | Cryo-EM structure of GPR30-Gq complex structure in the presence of fulvestrant | | 分子名称: | G-protein coupled estrogen receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Liu, H, Xu, P, Xu, H.E. | | 登録日 | 2024-01-01 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural and functional evidence that GPR30 is not a direct estrogen receptor.
Cell Res., 34, 2024
|
|
8XOG
 
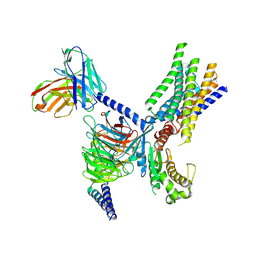 | | Cryo-EM structure of apo-GPR30-Gq complex structure | | 分子名称: | G-protein coupled estrogen receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Liu, H, Xu, P, Xu, H.E. | | 登録日 | 2024-01-01 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural and functional evidence that GPR30 is not a direct estrogen receptor.
Cell Res., 34, 2024
|
|
8XOF
 
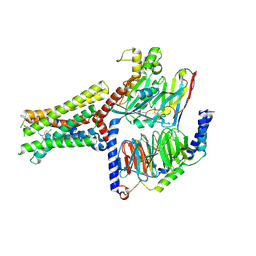 | | Cryo-EM structure of Lys05 bound GPR30-Gq complex structure | | 分子名称: | G protein subunit q, G-protein coupled estrogen receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Liu, H, Xu, P, Xu, H.E. | | 登録日 | 2024-01-01 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Structural and functional evidence that GPR30 is not a direct estrogen receptor.
Cell Res., 34, 2024
|
|
8HQS
 
 | | Cryo-EM structure of 8-subunit Smc5/6 head region | | 分子名称: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 3, Non-structural maintenance of chromosome element 4, ... | | 著者 | Qian, L, Jun, Z, Xiang, Z, Tong, C, Wang, Z, Duo, J, Zhenguo, C, Wang, L. | | 登録日 | 2022-12-14 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5DG3
 
 | |
8HTV
 
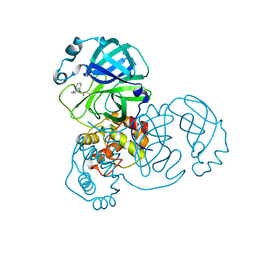 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 3a | | 分子名称: | 1-(5,6-dihydrobenzo[b][1]benzazepin-11-yl)-2-sulfanyl-ethanone, 3C-like proteinase | | 著者 | Su, H.X, Nie, T.Q, Li, M.J, Xu, Y.C. | | 登録日 | 2022-12-21 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Discovery and Mechanism Study of SARS-CoV-2 3C-like Protease Inhibitors with a New Reactive Group.
J.Med.Chem., 66, 2023
|
|
8I4U
 
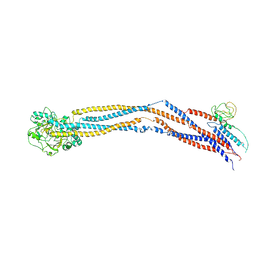 | | Cryo-EM structure of 5-subunit Smc5/6 hinge region | | 分子名称: | E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, Structural maintenance of chromosomes protein 6 | | 著者 | Qian, L, Jun, Z, Xiang, Z, Wang, Z, Tong, C, Duo, J, Zhenguo, C, Wang, L. | | 登録日 | 2023-01-21 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (6.73 Å) | | 主引用文献 | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8I4V
 
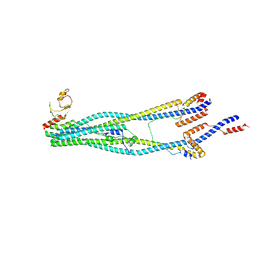 | | Cryo-EM structure of 5-subunit Smc5/6 arm region | | 分子名称: | DNA repair protein KRE29, E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, ... | | 著者 | Qian, L, Jun, Z, Xiang, Z, Cheng, T, Zhaoning, W, Zhenguo, C, Wang, L. | | 登録日 | 2023-01-21 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (5.97 Å) | | 主引用文献 | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8I13
 
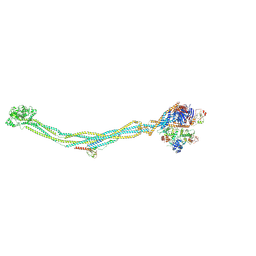 | | Cryo-EM structure of 6-subunit Smc5/6 | | 分子名称: | MMS21 isoform 1, NSE3 isoform 1, Non-structural maintenance of chromosomes element 1 homolog, ... | | 著者 | Qian, L, Jun, Z, Xiang, Z, Cheng, T, Zhaoning, W, Duo, J, Zhenguo, C, Wang, L. | | 登録日 | 2023-01-12 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (6.9 Å) | | 主引用文献 | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8I4W
 
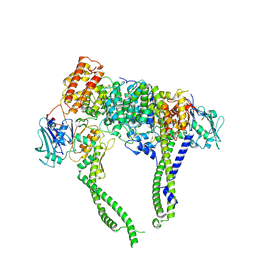 | | Cryo-EM structure of 5-subunit Smc5/6 head region | | 分子名称: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 5, Structural maintenance of chromosomes protein 5, ... | | 著者 | Qian, L, Jun, Z, Xiang, Z, Zhaoning, W, Cheng, T, Duo, J, Zhenguo, C, Wang, L. | | 登録日 | 2023-01-21 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (6.01 Å) | | 主引用文献 | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8I4X
 
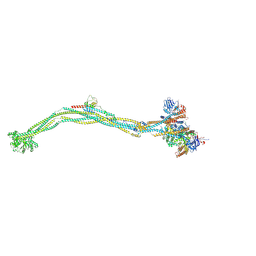 | | Cryo-EM structure of 5-subunit Smc5/6 | | 分子名称: | E3 SUMO-protein ligase MMS21, Non-structural maintenance of chromosome element 5, Nse6, ... | | 著者 | Qian, L, Jun, Z, Xiang, Z, Zhaoning, W, Tong, C, Duo, J, Zhenguo, C, Wang, L. | | 登録日 | 2023-01-21 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (8.5 Å) | | 主引用文献 | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8I21
 
 | | Cryo-EM structure of 6-subunit Smc5/6 arm region | | 分子名称: | E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, Structural maintenance of chromosomes protein 6 | | 著者 | Jun, Z, Qian, L, Xiang, Z, Tong, C, Zhaoning, W, Duo, J, Zhenguo, C, Lanfeng, W. | | 登録日 | 2023-01-13 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (6.02 Å) | | 主引用文献 | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5EMY
 
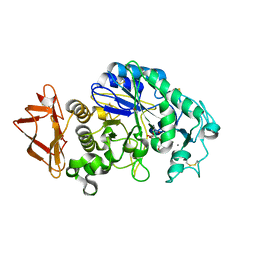 | | Human Pancreatic Alpha-Amylase in complex with the mechanism based inactivator glucosyl epi-cyclophellitol | | 分子名称: | (1R,2R,3S,5R,6S)-2,3,5-trihydroxy-6-(hydroxymethyl)cyclohexyl alpha-D-glucopyranoside, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Caner, S, Brayer, G.D. | | 登録日 | 2015-11-06 | | 公開日 | 2016-07-06 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.231 Å) | | 主引用文献 | Glucosyl epi-cyclophellitol allows mechanism-based inactivation and structural analysis of human pancreatic alpha-amylase.
Febs Lett., 590, 2016
|
|
2NOG
 
 | |
5GN8
 
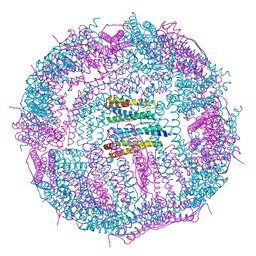 | | Structure of a 48-mer protein nanocage fabricated from its 24-mer analogue by subunit interface redesign | | 分子名称: | CALCIUM ION, Ferritin heavy chain | | 著者 | Zhang, S, Zang, J, Zhao, G, Mikami, B. | | 登録日 | 2016-07-19 | | 公開日 | 2016-12-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.805 Å) | | 主引用文献 | "Silent" Amino Acid Residues at Key Subunit Interfaces Regulate the Geometry of Protein Nanocages
ACS Nano, 10, 2016
|
|
4EMP
 
 | | Crystal structure of the mutant of ClpP E137A from Staphylococcus aureus | | 分子名称: | ATP-dependent Clp protease proteolytic subunit | | 著者 | Ye, F, Zhang, J, Liu, H, Luo, C, Yang, C.-G. | | 登録日 | 2012-04-12 | | 公開日 | 2013-04-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Helix unfolding/refolding characterizes the functional dynamics of Staphylococcus aureus Clp protease
J.Biol.Chem., 288, 2013
|
|
1AQL
 
 | |
2HNA
 
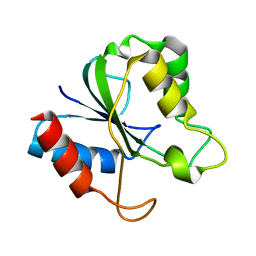 | | Solution Structure of a bacterial apo-flavodoxin | | 分子名称: | Protein mioC | | 著者 | Hu, Y, Jin, C. | | 登録日 | 2006-07-12 | | 公開日 | 2006-09-19 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structures and backbone dynamics of a flavodoxin MioC from Escherichia coli in both Apo- and Holo-forms: implications for cofactor binding and electron transfer
J.Biol.Chem., 281, 2006
|
|
2JSO
 
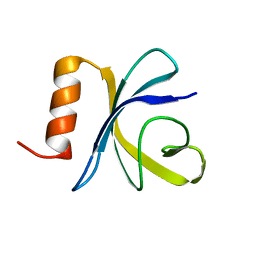 | | Antimicrobial resistance protein | | 分子名称: | Polymyxin resistance protein pmrD | | 著者 | Jin, C, Fu, W. | | 登録日 | 2007-07-10 | | 公開日 | 2007-09-04 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | First structure of the polymyxin resistance proteins.
Biochem.Biophys.Res.Commun., 361, 2007
|
|
1AKN
 
 | | STRUCTURE OF BILE-SALT ACTIVATED LIPASE | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILE-SALT ACTIVATED LIPASE | | 著者 | Wang, X, Zhang, X. | | 登録日 | 1997-05-23 | | 公開日 | 1998-05-27 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The crystal structure of bovine bile salt activated lipase: insights into the bile salt activation mechanism.
Structure, 5, 1997
|
|
2FKH
 
 | |
220L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BENZENE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
225L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BETA-MERCAPTOETHANOL, PARA-XYLENE, T4 LYSOZYME | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
228L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
226L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
