8JLO
 
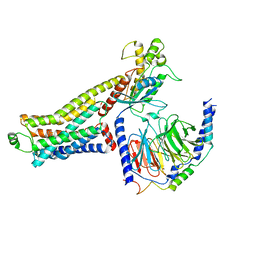 | | Ulotaront(SEP-363856)-bound hTAAR1-Gs protein complex | | 分子名称: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | 登録日 | 2023-06-02 | | 公開日 | 2023-11-15 | | 最終更新日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8JLR
 
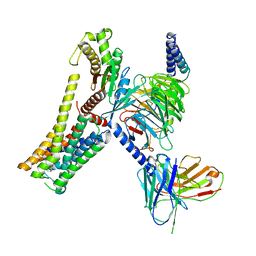 | | A77636-bound hTAAR1-Gs protein complex | | 分子名称: | (1~{S},3~{R})-3-(1-adamantyl)-1-(aminomethyl)-3,4-dihydro-1~{H}-isochromene-5,6-diol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | 登録日 | 2023-06-02 | | 公開日 | 2023-11-15 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8JLJ
 
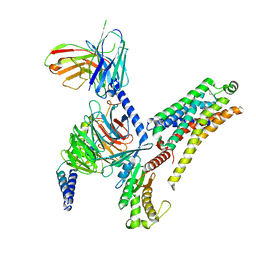 | | T1AM-bound mTAAR1-Gs protein complex | | 分子名称: | 4-[4-(2-azanylethyl)-2-iodanyl-phenoxy]phenol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | 登録日 | 2023-06-02 | | 公開日 | 2023-11-15 | | 最終更新日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
6M6I
 
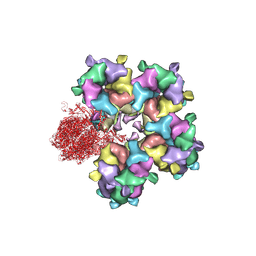 | | Structure of HSV2 B-capsid portal vertex | | 分子名称: | Coiled coils chain 1, Coiled coils chain 2, Major capsid protein, ... | | 著者 | Wang, X.X, Wang, N. | | 登録日 | 2020-03-14 | | 公開日 | 2021-03-10 | | 実験手法 | ELECTRON MICROSCOPY (4.05 Å) | | 主引用文献 | Structures of the portal vertex reveal essential protein-protein interactions for Herpesvirus assembly and maturation.
Protein Cell, 11, 2020
|
|
6M6H
 
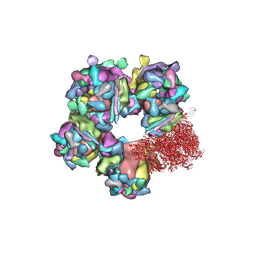 | | Structure of HSV2 C-capsid portal vertex | | 分子名称: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | 著者 | Wang, X.X, Wang, N. | | 登録日 | 2020-03-14 | | 公開日 | 2021-03-24 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structures of the portal vertex reveal essential protein-protein interactions for Herpesvirus assembly and maturation.
Protein Cell, 11, 2020
|
|
6M6G
 
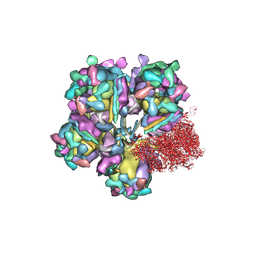 | | Structure of HSV2 viron capsid portal vertex | | 分子名称: | Capsid vertex component 1, Capsid vertex component 2, Coiled coils, ... | | 著者 | Wang, X.X, Wang, N. | | 登録日 | 2020-03-14 | | 公開日 | 2021-03-24 | | 実験手法 | ELECTRON MICROSCOPY (5.39 Å) | | 主引用文献 | Structures of the portal vertex reveal essential protein-protein interactions for Herpesvirus assembly and maturation.
Protein Cell, 11, 2020
|
|
6IPP
 
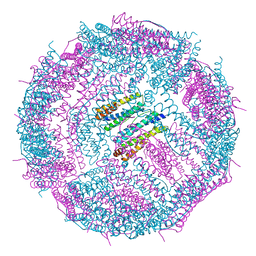 | | Non-native ferritin 8-mer mutant-C90A/C102A/C130A/D144C | | 分子名称: | FE (III) ION, Ferritin heavy chain | | 著者 | Zang, J, Chen, H, Zhou, K, Zhao, G. | | 登録日 | 2018-11-03 | | 公開日 | 2019-03-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.699 Å) | | 主引用文献 | Disulfide-mediated conversion of 8-mer bowl-like protein architecture into three different nanocages.
Nat Commun, 10, 2019
|
|
8K31
 
 | | The complex of WRKY33 C terminal DBD and SIB1 | | 分子名称: | Probable WRKY transcription factor 33, Sigma factor binding protein 1, chloroplastic, ... | | 著者 | Dong, X, Gong, Z, Hu, Y.F. | | 登録日 | 2023-07-14 | | 公開日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for the regulation of plant transcription factor WRKY33 by the VQ protein SIB1.
Commun Biol, 7, 2024
|
|
6IF4
 
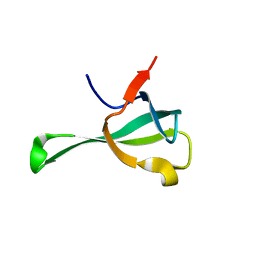 | | Crystal structure of Tbtudor | | 分子名称: | Histone acetyltransferase | | 著者 | Gao, J, Ye, K, Diwu, Y, Liao, S, Tu, X. | | 登録日 | 2018-09-18 | | 公開日 | 2019-09-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.934 Å) | | 主引用文献 | Crystal structure of TbEsa1 presumed Tudor domain from Trypanosoma brucei.
J.Struct.Biol., 209, 2020
|
|
3NMU
 
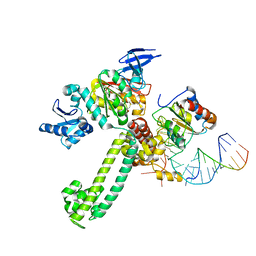 | | Crystal Structure of substrate-bound halfmer box C/D RNP | | 分子名称: | 50S ribosomal protein L7Ae, Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase, NOP5/NOP56 related protein, ... | | 著者 | Li, H, Xue, S, Wang, R. | | 登録日 | 2010-06-22 | | 公開日 | 2011-05-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.729 Å) | | 主引用文献 | Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle.
Mol.Cell, 39, 2010
|
|
2HNA
 
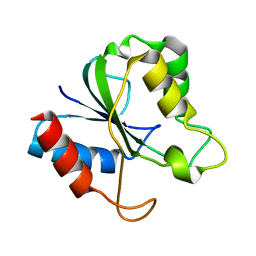 | | Solution Structure of a bacterial apo-flavodoxin | | 分子名称: | Protein mioC | | 著者 | Hu, Y, Jin, C. | | 登録日 | 2006-07-12 | | 公開日 | 2006-09-19 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structures and backbone dynamics of a flavodoxin MioC from Escherichia coli in both Apo- and Holo-forms: implications for cofactor binding and electron transfer
J.Biol.Chem., 281, 2006
|
|
3NBV
 
 | |
2FDV
 
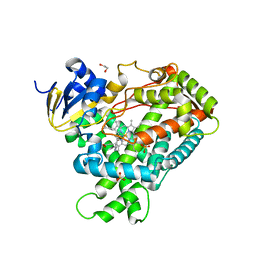 | | Microsomal P450 2A6 with the inhibitor N-Methyl(5-(pyridin-3-yl)furan-2-yl)methanamine bound | | 分子名称: | 1,2-ETHANEDIOL, Cytochrome P450 2A6, N-METHYL(5-(PYRIDIN-3-YL)FURAN-2-YL)METHANAMINE, ... | | 著者 | Yano, J.K, Stout, C.D, Johnson, E.F. | | 登録日 | 2005-12-14 | | 公開日 | 2006-11-28 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Synthetic Inhibitors of Cytochrome P-450 2A6: Inhibitory Activity, Difference Spectra, Mechanism of Inhibition, and Protein Cocrystallization.
J.Med.Chem., 49, 2006
|
|
5OSG
 
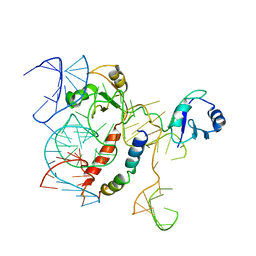 | | Structure of KSRP in context of Leishmania donovani 80S | | 分子名称: | 18S rRNA, 40S ribosomal protein S6, RNA binding protein, ... | | 著者 | Brito Querido, J, Mancera-Martinez, E, Vicens, Q, Bochler, A, Chicher, J, Simonetti, A, Hashem, Y. | | 登録日 | 2017-08-17 | | 公開日 | 2017-11-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | The cryo-EM Structure of a Novel 40S Kinetoplastid-Specific Ribosomal Protein.
Structure, 25, 2017
|
|
8F6H
 
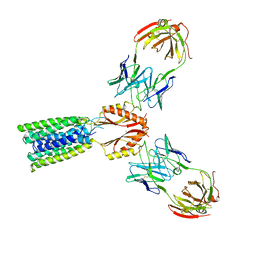 | | Cryo-EM structure of a Zinc-loaded asymmetrical TMD D70A mutant of the YiiP-Fab complex | | 分子名称: | Cadmium and zinc efflux pump FieF, Fab2r heavy chain, Fab2r light chain, ... | | 著者 | Lopez-Redondo, M.L, Hussein, A.K, Stokes, D.L. | | 登録日 | 2022-11-16 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Energy coupling and stoichiometry of Zn 2+ /H + antiport by the prokaryotic cation diffusion facilitator YiiP.
Elife, 12, 2023
|
|
2FDY
 
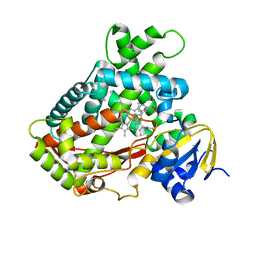 | | Microsomal P450 2A6 with the inhibitor Adrithiol bound | | 分子名称: | 4,4'-DIPYRIDYL DISULFIDE, Cytochrome P450 2A6, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Yano, J.K, Stout, C.D, Johnson, E.F. | | 登録日 | 2005-12-14 | | 公開日 | 2006-11-28 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Synthetic Inhibitors of Cytochrome P-450 2A6: Inhibitory Activity, Difference Spectra, Mechanism of Inhibition, and Protein Cocrystallization.
J.Med.Chem., 49, 2006
|
|
8F6E
 
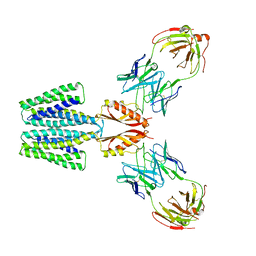 | | Cryo-EM structure of a Zinc-loaded wild-type YiiP-Fab complex | | 分子名称: | Cadmium and zinc efflux pump FieF, Fab heavy chain, Fab light chain, ... | | 著者 | Lopez-Redondo, M.L, Hussein, A.K, Stokes, D.L. | | 登録日 | 2022-11-16 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-08-21 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Energy coupling and stoichiometry of Zn 2+ /H + antiport by the prokaryotic cation diffusion facilitator YiiP.
Elife, 12, 2023
|
|
8F6F
 
 | | Cryo-EM structure of a Zinc-loaded D51A mutant of the YiiP-Fab complex | | 分子名称: | Cadmium and zinc efflux pump FieF, Fab2r heavy chain, Fab2r light chain, ... | | 著者 | Lopez-Redondo, M.L, Hussein, A.K, Stokes, D.L. | | 登録日 | 2022-11-16 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-08-21 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Energy coupling and stoichiometry of Zn 2+ /H + antiport by the prokaryotic cation diffusion facilitator YiiP.
Elife, 12, 2023
|
|
8F6K
 
 | |
8F6I
 
 | | Cryo-EM structure of a Zinc-loaded symmetrical D70A mutant of the YiiP-Fab complex | | 分子名称: | Cadmium and zinc efflux pump FieF, Fab2r heavy chain, Fab2r light chain, ... | | 著者 | Lopez-Redondo, M.L, Hussein, A.K, Stokes, D.L. | | 登録日 | 2022-11-16 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-08-21 | | 実験手法 | ELECTRON MICROSCOPY (4.03 Å) | | 主引用文献 | Energy coupling and stoichiometry of Zn 2+ /H + antiport by the prokaryotic cation diffusion facilitator YiiP.
Elife, 12, 2023
|
|
8F6J
 
 | | Cryo-EM structure of a Zinc-loaded D287A mutant of the YiiP-Fab complex | | 分子名称: | Cadmium and zinc efflux pump FieF, Fab2r heavy chain, Fab2r light chain, ... | | 著者 | Lopez-Redondo, M.L, Hussein, A.K, Stokes, D.L. | | 登録日 | 2022-11-16 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-08-21 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Energy coupling and stoichiometry of Zn 2+ /H + antiport by the prokaryotic cation diffusion facilitator YiiP.
Elife, 12, 2023
|
|
3GXB
 
 | | Crystal structure of VWF A2 domain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, ... | | 著者 | Zhou, Y.F, Springer, T.A. | | 登録日 | 2009-04-02 | | 公開日 | 2009-05-05 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural specializations of A2, a force-sensing domain in the ultralarge vascular protein von Willebrand factor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2HNB
 
 | | Solution Structure of a bacterial holo-flavodoxin | | 分子名称: | Protein mioC | | 著者 | Hu, Y, Jin, C. | | 登録日 | 2006-07-12 | | 公開日 | 2006-09-19 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structures and backbone dynamics of a flavodoxin MioC from Escherichia coli in both Apo- and Holo-forms: implications for cofactor binding and electron transfer
J.Biol.Chem., 281, 2006
|
|
7KX5
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with noncovalent inhibitor Jun8-76-3A | | 分子名称: | 3C-like proteinase, GLYCEROL, N-([1,1'-biphenyl]-4-yl)-N-[(1R)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]furan-2-carboxamide | | 著者 | Sacco, M, Wang, J, Chen, Y. | | 登録日 | 2020-12-03 | | 公開日 | 2020-12-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Discovery of Di- and Trihaloacetamides as Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity.
J.Am.Chem.Soc., 143, 2021
|
|
6LQJ
 
 | | Low resolution architecture of curli complex | | 分子名称: | Curli production assembly/transport component CsgF, Curli production assembly/transport component CsgG | | 著者 | Zhang, M, Shi, H, Huang, Y. | | 登録日 | 2020-01-13 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.24 Å) | | 主引用文献 | Cryo-EM structure of the nonameric CsgG-CsgF complex and its implications for controlling curli biogenesis in Enterobacteriaceae.
Plos Biol., 18, 2020
|
|
