8ZOZ
 
 | | Crystal structure of the complex of glyceraldehyde-3-phosphate dehydrogenase of type B from Acinetobacter baumannii with Adenosine monophosphate at 3.20 A resolution. | | 分子名称: | ADENOSINE MONOPHOSPHATE, Glyceraldehyde-3-phosphate dehydrogenase, SULFATE ION | | 著者 | Pahuja, P, Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Sharma, S, Raje, C.I, Singh, T.P. | | 登録日 | 2024-05-29 | | 公開日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Crystal structure of the complex of glyceraldehyde-3-phosphate dehydrogenase of type B from Acinetobacter baumannii with Adenosine monophosphate at 3.20 A resolution.
To Be Published
|
|
6JBP
 
 | | Structure of MP-4 from Mucuna pruriens at 2.22 Angstroms | | 分子名称: | Kunitz-type trypsin inhibitor-like 2 protein | | 著者 | Jain, A, Shikhi, M, Kumar, A, Kumar, A, Nair, D.T, Salunke, D.M. | | 登録日 | 2019-01-26 | | 公開日 | 2020-01-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.217 Å) | | 主引用文献 | The structure of MP-4 from Mucuna pruriens at 2.22 angstrom resolution.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
8WT1
 
 | | Crystal structure of S9 carboxypeptidase from Geobacillus sterothermophilus | | 分子名称: | ALANINE, CITRATE ANION, GLYCEROL, ... | | 著者 | Chandravanshi, K, Kumar, A, Sen, C, Singh, R, Bhange, G.B, Makde, R.D. | | 登録日 | 2023-10-17 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure and solution scattering of Geobacillus stearothermophilus S9 peptidase reveal structural adaptations for carboxypeptidase activity.
Febs Lett., 598, 2024
|
|
7F8S
 
 | | Pennisetum glaucum (Pearl millet) dehydroascorbate reductase (DHAR) with catalytic cysteine (Cy20) in sulphenic and sulfinic acid forms. | | 分子名称: | Dehydroascorbate reductase, SULFATE ION | | 著者 | Das, B.K, Kumar, A, Sreeshma, N.S, Arockiasamy, A. | | 登録日 | 2021-07-02 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | Comparative kinetic analysis of ascorbate (Vitamin-C) recycling dehydroascorbate reductases from plants and humans.
Biochem.Biophys.Res.Commun., 591, 2021
|
|
1BPD
 
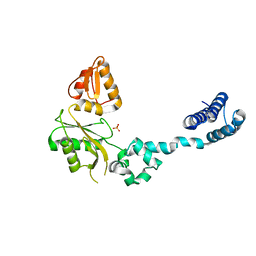 | | CRYSTAL STRUCTURE OF RAT DNA POLYMERASE BETA: EVIDENCE FOR A COMMON POLYMERASE MECHANISM | | 分子名称: | DNA POLYMERASE BETA, PHOSPHATE ION | | 著者 | Sawaya, M.R, Pelletier, H, Kumar, A, Wilson, S.H, Kraut, J. | | 登録日 | 1994-04-12 | | 公開日 | 1994-06-22 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism.
Science, 264, 1994
|
|
1BPB
 
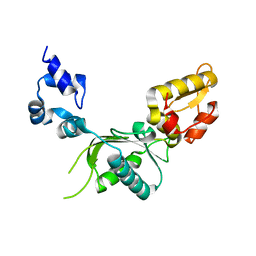 | | CRYSTAL STRUCTURE OF RAT DNA POLYMERASE BETA: EVIDENCE FOR A COMMON POLYMERASE MECHANISM | | 分子名称: | DNA POLYMERASE BETA | | 著者 | Sawaya, M.R, Pelletier, H, Kumar, A, Wilson, S.H, Kraut, J. | | 登録日 | 1994-04-12 | | 公開日 | 1994-06-22 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism.
Science, 264, 1994
|
|
7Q4V
 
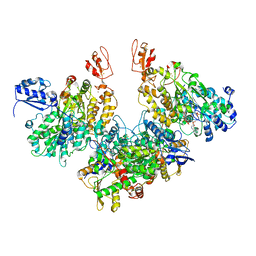 | | Electron bifurcating hydrogenase - HydABC from A. woodii | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | 著者 | Katsyv, A, Kumar, A, Saura, P, Poeverlein, M.C, Freibert, S.A, Stripp, S, Jain, S, Gamiz-Hernandez, A.P, Kaila, V.R.I, Mueller, V, Schuller, J.M. | | 登録日 | 2021-11-02 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-03-29 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Molecular Basis of the Electron Bifurcation Mechanism in the [FeFe]-Hydrogenase Complex HydABC.
J.Am.Chem.Soc., 145, 2023
|
|
8JWV
 
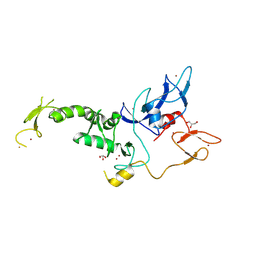 | | Untethered R0RBR | | 分子名称: | BARIUM ION, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | 著者 | Lenka, D.R, Kumar, A. | | 登録日 | 2023-06-29 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-09-18 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Additional feedforward mechanism of Parkin activation via binding of phospho-UBL and RING0 in trans.
Elife, 13, 2024
|
|
7XQ7
 
 | | The complex structure of WT-Mpro | | 分子名称: | 3C-like proteinase nsp5, SODIUM ION | | 著者 | Sahoo, P, Lenka, D.R, Kumar, A. | | 登録日 | 2022-05-06 | | 公開日 | 2023-03-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Detailed Insights into the Inhibitory Mechanism of New Ebselen Derivatives against Main Protease (M pro ) of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2).
Acs Pharmacol Transl Sci, 6, 2023
|
|
7XQ6
 
 | | The complex structure of mutant Mpro with inhibitor | | 分子名称: | 3C-like proteinase nsp5, CHLORIDE ION | | 著者 | Sahoo, P, Lenka, D.R, Kumar, A. | | 登録日 | 2022-05-06 | | 公開日 | 2023-03-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Detailed Insights into the Inhibitory Mechanism of New Ebselen Derivatives against Main Protease (M pro ) of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2).
Acs Pharmacol Transl Sci, 6, 2023
|
|
8V8K
 
 | | Crystal Structure of Nanobody NbE | | 分子名称: | Nanobody NbE | | 著者 | Koehl, A, Manglik, A, Yu, J, Kumar, A, Zhang, X, Martin, C, Raia, P, Steyaert, J, Ballet, S, Boland, A, Stoeber, M. | | 登録日 | 2023-12-05 | | 公開日 | 2024-09-11 | | 最終更新日 | 2024-10-02 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structural Basis of mu-Opioid Receptor-Targeting by a Nanobody Antagonist
To Be Published
|
|
6IFG
 
 | | Crystal structure of M1 zinc metallopeptidase E323A mutant bound to Tyr-ser-ala substrate from Deinococcus radiodurans | | 分子名称: | FORMIC ACID, Tripeptides (TYR-SER-ALA), ZINC ION, ... | | 著者 | Agrawal, R, Kumar, A, Kumar, A, Makde, R.D. | | 登録日 | 2018-09-20 | | 公開日 | 2019-09-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Two-domain aminopeptidase of M1 family: Structural features for substrate binding and gating in absence of C-terminal domain.
J.Struct.Biol., 208, 2019
|
|
7B7F
 
 | | Room temperature X-ray structure of H/D-exchanged PLL lectin in complex with L-fucose | | 分子名称: | PLL lectin, alpha-L-fucopyranose, beta-L-fucopyranose | | 著者 | Gajdos, L, Blakeley, M.P, Kumar, A, Wimmerova, M, Haertlein, M, Forsyth, V.T, Imberty, A, Devos, J.M. | | 登録日 | 2020-12-10 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Visualization of hydrogen atoms in a perdeuterated lectin-fucose complex reveals key details of protein-carbohydrate interactions.
Structure, 29, 2021
|
|
7B7E
 
 | | Room temperature X-ray structure of perdeuterated PLL lectin | | 分子名称: | PLL lectin | | 著者 | Gajdos, L, Blakeley, M.P, Kumar, A, Wimmerova, M, Haertlein, M, Forsyth, V.T, Imberty, A, Devos, J.M. | | 登録日 | 2020-12-10 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Visualization of hydrogen atoms in a perdeuterated lectin-fucose complex reveals key details of protein-carbohydrate interactions.
Structure, 29, 2021
|
|
7B7C
 
 | | Room temperature X-ray structure of perdeuterated PLL lectin in complex with L-fucose | | 分子名称: | PLL lectin, alpha-L-fucopyranose, beta-L-fucopyranose | | 著者 | Gajdos, L, Blakeley, M.P, Kumar, A, Wimmerova, M, Haertlein, M, Forsyth, V.T, Imberty, A, Devos, J.M. | | 登録日 | 2020-12-10 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Visualization of hydrogen atoms in a perdeuterated lectin-fucose complex reveals key details of protein-carbohydrate interactions.
Structure, 29, 2021
|
|
6IFF
 
 | | Crystal structure of M1 zinc metallopeptidase E323A mutant from Deinococcus radiodurans | | 分子名称: | SODIUM ION, TYROSINE, ZINC ION, ... | | 著者 | Agrawal, R, Kumar, A, Kumar, A, Gaur, N.K, Makde, R.D. | | 登録日 | 2018-09-20 | | 公開日 | 2019-09-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Structural basis for the unusual substrate specificity of unique two-domain M1 metallopeptidase.
Int.J.Biol.Macromol., 147, 2020
|
|
7BBI
 
 | | Joint X-ray/neutron room temperature structure of H/D-exchanged PLL lectin | | 分子名称: | PLL lectin | | 著者 | Gajdos, L, Blakeley, M.P, Kumar, A, Wimmerova, M, Haertlein, M, Forsyth, V.T, Imberty, A, Devos, J.M. | | 登録日 | 2020-12-17 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-10-09 | | 実験手法 | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | 主引用文献 | Visualization of hydrogen atoms in a perdeuterated lectin-fucose complex reveals key details of protein-carbohydrate interactions.
Structure, 29, 2021
|
|
7BB4
 
 | | Crystal structure of perdeuterated PLL lectin in complex with L-fucose | | 分子名称: | GLYCEROL, PLL lectin, alpha-L-fucopyranose, ... | | 著者 | Gajdos, L, Blakeley, M.P, Kumar, A, Wimmerova, M, Haertlein, M, Forsyth, V.T, Imberty, A, Devos, J.M. | | 登録日 | 2020-12-16 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Visualization of hydrogen atoms in a perdeuterated lectin-fucose complex reveals key details of protein-carbohydrate interactions.
Structure, 29, 2021
|
|
7BBC
 
 | | Joint X-ray/neutron room temperature structure of perdeuterated PLL lectin in complex with perdeuterated L-fucose | | 分子名称: | PLL lectin, alpha-L-fucopyranose, beta-L-fucopyranose | | 著者 | Gajdos, L, Blakeley, M.P, Kumar, A, Wimmerova, M, Haertlein, M, Forsyth, V.T, Imberty, A, Devos, J.M. | | 登録日 | 2020-12-17 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-10-09 | | 実験手法 | NEUTRON DIFFRACTION (1.84 Å), X-RAY DIFFRACTION | | 主引用文献 | Visualization of hydrogen atoms in a perdeuterated lectin-fucose complex reveals key details of protein-carbohydrate interactions.
Structure, 29, 2021
|
|
6EOJ
 
 | | PolyA polymerase module of the cleavage and polyadenylation factor (CPF) from Saccharomyces cerevisiae | | 分子名称: | Polyadenylation factor subunit 2,Polyadenylation factor subunit 2, Protein CFT1, ZINC ION, ... | | 著者 | Casanal, A, Kumar, A, Hill, C.H, Emsley, P, Passmore, L.A. | | 登録日 | 2017-10-09 | | 公開日 | 2017-11-15 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.55 Å) | | 主引用文献 | Architecture of eukaryotic mRNA 3'-end processing machinery.
Science, 358, 2017
|
|
8IYO
 
 | | Crystal structure of a protein acetyltransferase, HP0935, acetyl-CoA bound form | | 分子名称: | ACETYL COENZYME *A, N-acetyltransferase domain-containing protein | | 著者 | Dadireddy, V, Mahanta, P, Kumar, A, Desirazu, R.N, Ramakumar, S. | | 登録日 | 2023-04-05 | | 公開日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of a protein acetyltransferase, HP0935, acetyl-CoA bound form
To be published
|
|
8IYM
 
 | | Crystal structure of a protein acetyltransferase, HP0935 | | 分子名称: | 1,2-ETHANEDIOL, N-acetyltransferase domain-containing protein, POTASSIUM ION, ... | | 著者 | Dadireddy, V, Mahanta, P, Kumar, A, Desirazu, R.N, Ramakumar, S. | | 登録日 | 2023-04-05 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of a protein acetyltransferase, HP0935
To be published
|
|
6IDN
 
 | | Crystal structure of ICChI chitinase from ipomoea carnea | | 分子名称: | CALCIUM ION, ICChI, a glycosylated chitinase, ... | | 著者 | Kumar, S, Kumar, A, Patel, A.K. | | 登録日 | 2018-09-10 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | TIM barrel fold and glycan moieties in the structure of ICChI, a protein with chitinase and lysozyme activity.
Phytochemistry, 170, 2020
|
|
5HWK
 
 | | Crystal structure of gama glutamyl cyclotransferease specific to glutathione from yeast | | 分子名称: | BENZOIC ACID, Glutathione-specific gamma-glutamylcyclotransferase, PHOSPHATE ION | | 著者 | Kaur, A, Gautam, R, Srivastava, R, Chandel, A, Kumar, A, Karthikeyan, S, Bachhawat, A.K. | | 登録日 | 2016-01-29 | | 公開日 | 2016-12-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.344 Å) | | 主引用文献 | ChaC2, an Enzyme for Slow Turnover of Cytosolic Glutathione
J. Biol. Chem., 292, 2017
|
|
5HWI
 
 | | Crystal structure of selenomethionine labelled gama glutamyl cyclotransferease specific to glutathione from yeast | | 分子名称: | GLYCEROL, Glutathione-specific gamma-glutamylcyclotransferase, SUCCINIC ACID | | 著者 | Kaur, A, Gautam, R, Srivastava, R, Chandel, A, Kumar, A, Karthikeyan, S, Bachhawat, A.K. | | 登録日 | 2016-01-29 | | 公開日 | 2016-12-14 | | 最終更新日 | 2017-01-25 | | 実験手法 | X-RAY DIFFRACTION (1.755 Å) | | 主引用文献 | ChaC2, an Enzyme for Slow Turnover of Cytosolic Glutathione
J. Biol. Chem., 292, 2017
|
|
