3WN6
 
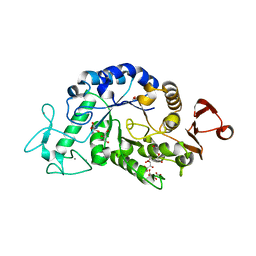 | | Crystal structure of alpha-amylase AmyI-1 from Oryza sativa | | 分子名称: | Alpha-amylase, CALCIUM ION, D(-)-TARTARIC ACID, ... | | 著者 | Ochiai, A, Sugai, H, Harada, K, Tanaka, S, Ishiyama, Y, Ito, K, Tanaka, T, Uchiumi, T, Taniguchi, M, Mitsui, T. | | 登録日 | 2013-12-05 | | 公開日 | 2014-09-10 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Crystal structure of alpha-amylase from Oryza sativa: molecular insights into enzyme activity and thermostability
Biosci.Biotechnol.Biochem., 78, 2014
|
|
1J3A
 
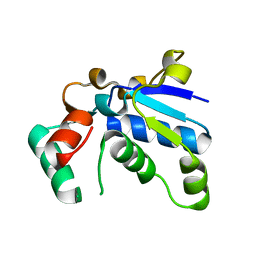 | | Crystal structure of ribosomal protein L13 from Pyrococcus horikoshii | | 分子名称: | 50S ribosomal protein L13P | | 著者 | Nakashima, T, Tanaka, M, Kazama, T, Kawamura, S, Kimura, M, Yao, M, Tanaka, I. | | 登録日 | 2003-01-21 | | 公開日 | 2003-02-04 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of ribosomal protein L13 from hyperthermophilic archaeon Pyrococcus horikoshii
To be Published
|
|
1DPT
 
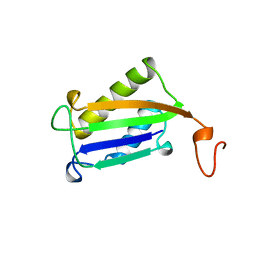 | | D-DOPACHROME TAUTOMERASE | | 分子名称: | D-DOPACHROME TAUTOMERASE | | 著者 | Sugimoto, H, Taniguchi, M, Nakagawa, A, Tanaka, I. | | 登録日 | 1998-05-11 | | 公開日 | 1999-03-30 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Crystal structure of human D-dopachrome tautomerase, a homologue of macrophage migration inhibitory factor, at 1.54 A resolution.
Biochemistry, 38, 1999
|
|
7ZAV
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3 | | 著者 | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | 登録日 | 2022-03-22 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZA1
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | 著者 | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | 登録日 | 2022-03-21 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (4.1 Å) | | 主引用文献 | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZA3
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | 著者 | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | 登録日 | 2022-03-21 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (4 Å) | | 主引用文献 | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZAW
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3 | | 著者 | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | 登録日 | 2022-03-22 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZA2
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | 著者 | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | 登録日 | 2022-03-21 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (4.6 Å) | | 主引用文献 | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
4YOX
 
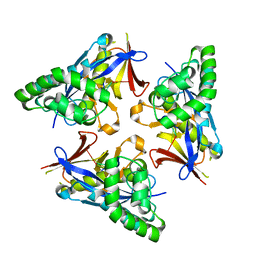 | |
4YOU
 
 | |
5MUD
 
 | | Crystal structure of an amyloidogenic light chain dimer H6 | | 分子名称: | light chain dimer,IGL@ protein | | 著者 | Oberti, L, Bacarizo, J, Maritan, M, Rognoni, P, Bolognesi, M, Ricagno, S. | | 登録日 | 2017-01-13 | | 公開日 | 2017-12-13 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
4YOR
 
 | |
3R8I
 
 | | Crystal Structure of PPARgamma with an achiral ureidofibrate derivative (RT86) | | 分子名称: | 2-(4-{2-[1,3-benzoxazol-2-yl(heptyl)amino]ethyl}phenoxy)-2-methylpropanoic acid, Peroxisome proliferator-activated receptor gamma | | 著者 | Pochetti, G, Montanari, R, Loiodice, F, Laghezza, A, Fracchiolla, G, Lavecchia, A, Novellino, E, Crestani, M. | | 登録日 | 2011-03-24 | | 公開日 | 2012-01-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Synthesis, characterization and biological evaluation of ureidofibrate-like derivatives endowed with peroxisome proliferator-activated receptor activity.
J.Med.Chem., 55, 2012
|
|
4YOY
 
 | |
5MUH
 
 | | Crystal structure of an amyloidogenic light chain dimer H7 | | 分子名称: | light chain dimer | | 著者 | Oberti, L, Rognoni, P, Russo, R, Maritan, M, Bacarizo, J, Bolognesi, M, Ricagno, S. | | 登録日 | 2017-01-13 | | 公開日 | 2017-12-13 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
4YOV
 
 | |
4YOW
 
 | |
6KFG
 
 | | Undocked INX-6 hemichannel in detergent | | 分子名称: | Innexin-6 | | 著者 | Burendei, B, Shinozaki, R, Watanabe, M, Terada, T, Tani, K, Fujiyoshi, Y, Oshima, A. | | 登録日 | 2019-07-07 | | 公開日 | 2020-02-12 | | 最終更新日 | 2020-03-11 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structures of undocked innexin-6 hemichannels in phospholipids.
Sci Adv, 6, 2020
|
|
4YOT
 
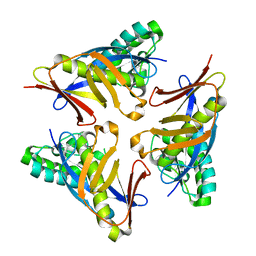 | |
6KFF
 
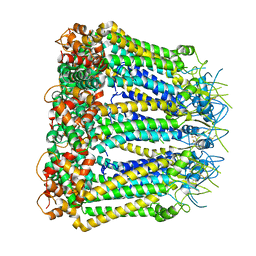 | | Undocked INX-6 hemichannel in a nanodisc | | 分子名称: | Innexin-6 | | 著者 | Burendei, B, Shinozaki, R, Watanabe, M, Terada, T, Tani, K, Fujiyoshi, Y, Oshima, A. | | 登録日 | 2019-07-07 | | 公開日 | 2020-02-12 | | 最終更新日 | 2020-03-11 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structures of undocked innexin-6 hemichannels in phospholipids.
Sci Adv, 6, 2020
|
|
6KFH
 
 | | Undocked hemichannel of an N-terminal deletion mutant of INX-6 in a nanodisc | | 分子名称: | Innexin-6 | | 著者 | Burendei, B, Shinozaki, R, Watanabe, M, Terada, T, Tani, K, Fujiyoshi, Y, Oshima, A. | | 登録日 | 2019-07-07 | | 公開日 | 2020-02-12 | | 最終更新日 | 2020-03-11 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM structures of undocked innexin-6 hemichannels in phospholipids.
Sci Adv, 6, 2020
|
|
5LGV
 
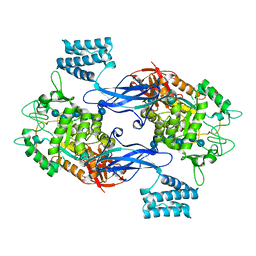 | | GlgE isoform 1 from Streptomyces coelicolor E423A mutant soaked in maltooctaose | | 分子名称: | Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Syson, K, Stevenson, C.E.M, Mia, F, Barclay, J.E, Tang, M, Gorelik, A, Rashid, A.M, Lawson, D.M, Bornemann, S. | | 登録日 | 2016-07-08 | | 公開日 | 2016-08-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Ligand-bound structures and site-directed mutagenesis identify the acceptor and secondary binding sites of Streptomyces coelicolor maltosyltransferase GlgE.
J.Biol.Chem., 291, 2016
|
|
5LGW
 
 | | GlgE isoform 1 from Streptomyces coelicolor D394A mutant co-crystallised with maltodextrin | | 分子名称: | 1,2-ETHANEDIOL, Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, CITRIC ACID, ... | | 著者 | Syson, K, Stevenson, C.E.M, Mia, F, Barclay, J.E, Tang, M, Gorelik, A, Rashid, A.M, Lawson, D.M, Bornemann, S. | | 登録日 | 2016-07-08 | | 公開日 | 2016-08-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Ligand-bound structures and site-directed mutagenesis identify the acceptor and secondary binding sites of Streptomyces coelicolor maltosyltransferase GlgE.
J.Biol.Chem., 291, 2016
|
|
6Q35
 
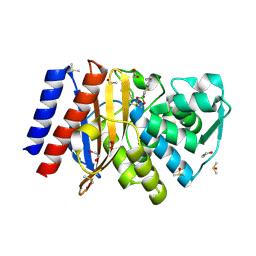 | | Crystal structure of GES-5 beta-lactamase in complex with boronic inhibitor cpd 3 | | 分子名称: | 1,2-ETHANEDIOL, Beta-lactamase, DIMETHYL SULFOXIDE, ... | | 著者 | Maso, L, Quotadamo, A, Bellio, P, Montanari, M, Venturelli, A, Celenza, G, Costi, M.P, Tondi, D, Cendron, L. | | 登録日 | 2018-12-03 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | X-ray Crystallography Deciphers the Activity of Broad-Spectrum Boronic Acid beta-Lactamase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6Q30
 
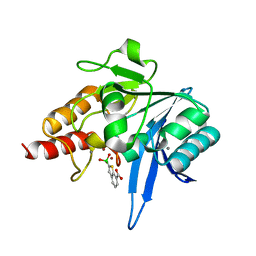 | | Crystal structure of NDM-1 beta-lactamase in complex with boronic inhibitor cpd 5 | | 分子名称: | (7-carboxy-1-benzothiophen-2-yl)-tris(oxidanyl)boranuide, CALCIUM ION, Metallo-beta-lactamase type 2, ... | | 著者 | Maso, L, Quotadamo, A, Bellio, P, Montanari, M, Celenza, G, Venturelli, A, Costi, M.P, Tondi, D, Cendron, L. | | 登録日 | 2018-12-03 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | X-ray Crystallography Deciphers the Activity of Broad-Spectrum Boronic Acid beta-Lactamase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
