1O66
 
 | |
1O60
 
 | |
1MDO
 
 | | Crystal structure of ArnB aminotransferase with pyridomine 5' phosphate | | 分子名称: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ArnB aminotransferase | | 著者 | Noland, B.W, Newman, J.M, Hendle, J, Badger, J, Christopher, J.A, Tresser, J, Buchanan, M.D, Wright, T, Rutter, M.E, Sanderson, W.E, Muller-Dieckmann, H.-J, Gajiwala, K, Sauder, J.M, Buchanan, S.G. | | 登録日 | 2002-08-07 | | 公開日 | 2002-12-11 | | 最終更新日 | 2018-12-26 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural studies of Salmonella typhimurium ArnB (PmrH) aminotransferase: A 4-amino-4-deoxy-L-arabinose lipopolysaccharide modifying enzyme
Structure, 10, 2002
|
|
1O67
 
 | |
1O61
 
 | |
1O69
 
 | | Crystal structure of a PLP-dependent enzyme | | 分子名称: | (2-AMINO-4-FORMYL-5-HYDROXY-6-METHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, BETA-MERCAPTOETHANOL, aminotransferase | | 著者 | Structural GenomiX | | 登録日 | 2003-10-23 | | 公開日 | 2003-11-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1O64
 
 | |
1O65
 
 | |
1O68
 
 | |
1O6D
 
 | |
1O62
 
 | |
1MPG
 
 | | 3-METHYLADENINE DNA GLYCOSYLASE II FROM ESCHERICHIA COLI | | 分子名称: | 3-METHYLADENINE DNA GLYCOSYLASE II, GLYCEROL | | 著者 | Labahn, J, Schaerer, O.D, Long, A, Ezaz-Nikpay, K, Verdine, G.L, Ellenberger, T.E. | | 登録日 | 1997-10-28 | | 公開日 | 1998-01-28 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for the excision repair of alkylation-damaged DNA.
Cell(Cambridge,Mass.), 86, 1996
|
|
1MDZ
 
 | | Crystal structure of ArnB aminotransferase with cycloserine and pyridoxal 5' phosphate | | 分子名称: | ArnB aminotransferase, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Noland, B.W, Newman, J.M, Hendle, J, Badger, J, Christopher, J.A, Tresser, J, Buchanan, M.D, Wright, T, Rutter, M.E, Sanderson, W.E, Muller-Dieckmann, H.-J, Gajiwala, K.S, Sauder, J.M, Buchanan, S.G. | | 登録日 | 2002-08-07 | | 公開日 | 2002-12-11 | | 最終更新日 | 2018-12-26 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Structural studies of Salmonella typhimurium ArnB (PmrH) aminotransferase: A 4-amino-4-deoxy-L-arabinose lipopolysaccharide modifying enzyme
Structure, 10, 2002
|
|
1O63
 
 | |
1O6C
 
 | |
2DDQ
 
 | |
4WUA
 
 | | Crystal structure of human SRPK1 complexed to an inhibitor SRPIN340 | | 分子名称: | CITRIC ACID, N-[2-(1-piperidinyl)-5-(trifluoromethyl)phenyl]-4-pyridinecarboxamide, SRSF protein kinase 1, ... | | 著者 | Hoshina, M, Ikura, T, Hosoya, T, Hagiwara, M, Ito, N. | | 登録日 | 2014-10-31 | | 公開日 | 2015-09-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Identification of a Dual Inhibitor of SRPK1 and CK2 That Attenuates Pathological Angiogenesis of Macular Degeneration in Mice
Mol.Pharmacol., 88, 2015
|
|
8HX6
 
 | |
8HX7
 
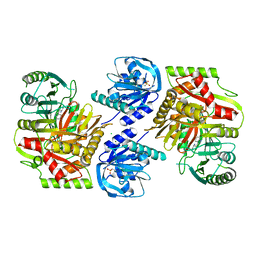 | |
8HX9
 
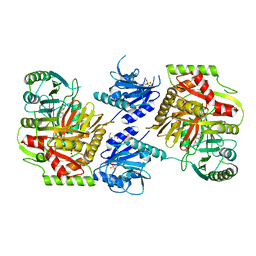 | | Crystal structure of 4-amino-4-deoxychorismate synthase from Streptomyces venezuelae with chorismate | | 分子名称: | (3R,4R)-3-[(1-carboxyethenyl)oxy]-4-hydroxycyclohexa-1,5-diene-1-carboxylic acid, 4-amino-4-deoxychorismate synthase, FORMIC ACID, ... | | 著者 | Nakamichi, Y, Watanabe, M. | | 登録日 | 2023-01-04 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Structural basis for the allosteric pathway of 4-amino-4-deoxychorismate synthase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
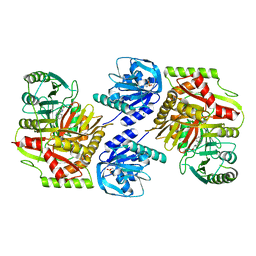
8HX8
 
 | |
4RU4
 
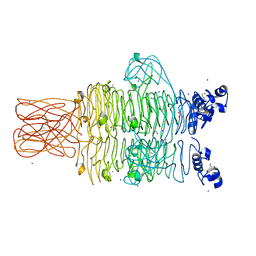 | | Crystal structure of the tailspike protein gp49 from Pseudomonas phage LKA1 | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, SODIUM ION, ... | | 著者 | Browning, C, Shneider, M.M, Leiman, P.G. | | 登録日 | 2014-11-18 | | 公開日 | 2015-11-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.903 Å) | | 主引用文献 | The O-specific polysaccharide lyase from the phage LKA1 tailspike reduces Pseudomonas virulence.
Sci Rep, 7, 2017
|
|
4RU5
 
 | | Crystal Structure of the Pseudomonas phage phi297 tailspike gp61 | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | 著者 | Browning, C, Sycheva, L.V, Shneider, M.M, Leiman, P.G. | | 登録日 | 2014-11-18 | | 公開日 | 2015-11-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | The O-specific polysaccharide lyase from the phage LKA1 tailspike reduces Pseudomonas virulence.
Sci Rep, 7, 2017
|
|
1VHN
 
 | |
1UGU
 
 | | Crystal structure of PYP E46Q mutant | | 分子名称: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | 著者 | Sugishima, M, Tanimoto, Y, Hamada, N, Tokunaga, F, Fukuyama, K. | | 登録日 | 2003-06-19 | | 公開日 | 2004-08-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structure of photoactive yellow protein (PYP) E46Q mutant at 1.2 A resolution suggests how Glu46 controls the spectroscopic and kinetic characteristics of PYP.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
