8SJV
 
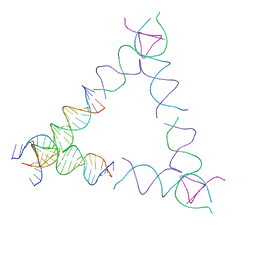 | | [4T24] Self-assembling left-handed tensegrity triangle with 24 interjunction base pairs and R3 symmetry | | 分子名称: | DNA (5'-D(P*CP*TP*TP*GP*TP*AP*GP*TP*CP*TP*CP*AP*CP*CP*AP*CP*TP*GP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(P*GP*AP*AP*CP*AP*CP*TP*CP*CP*TP*GP*AP*GP*AP*CP*TP*AP*CP*AP*A)-3'), DNA (5'-D(P*GP*AP*CP*AP*TP*CP*AP*CP*AP*GP*TP*GP*GP*AP*CP*TP*AP*CP*AP*AP*G)-3'), ... | | 著者 | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | 登録日 | 2023-04-18 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (8.59 Å) | | 主引用文献 | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6E1Z
 
 | |
6E23
 
 | |
6E22
 
 | |
6E1Y
 
 | |
6DYA
 
 | | WDR5 in complex with a WIN site inhibitor | | 分子名称: | DIMETHYL SULFOXIDE, N-[(3,5-dichlorophenyl)methyl]-3-[(1H-imidazol-1-yl)methyl]benzamide, SULFATE ION, ... | | 著者 | Phan, J, Wang, F, Fesik, S.W. | | 登録日 | 2018-07-01 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Displacement of WDR5 from Chromatin by a WIN Site Inhibitor with Picomolar Affinity.
Cell Rep, 26, 2019
|
|
6V2F
 
 | | Crystal structure of the HIV capsid hexamer bound to the small molecule long-acting inhibitor, GS-6207 | | 分子名称: | HIV-1 capsid, N-[(1S)-1-(3-{4-chloro-3-[(methylsulfonyl)amino]-1-(2,2,2-trifluoroethyl)-1H-indazol-7-yl}-6-[3-methyl-3-(methylsulfonyl)but-1-yn-1-yl]pyridin-2-yl)-2-(3,5-difluorophenyl)ethyl]-2-[(3bS,4aR)-5,5-difluoro-3-(trifluoromethyl)-3b,4,4a,5-tetrahydro-1H-cyclopropa[3,4]cyclopenta[1,2-c]pyrazol-1-yl]acetamide | | 著者 | Appleby, T.C, Link, J.O, Yant, S.R, Villasenor, A.G, Somoza, J.R, Hu, E.Y, Schroeder, S.D, Cihlar, T. | | 登録日 | 2019-11-22 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Clinical targeting of HIV capsid protein with a long-acting small molecule.
Nature, 584, 2020
|
|
6BGH
 
 | |
7E7X
 
 | |
7E7Y
 
 | |
7E8C
 
 | | SARS-CoV-2 S-6P in complex with 9 Fabs | | 分子名称: | 368-2 H, 368-2 L, 604 H, ... | | 著者 | Du, S, Xiao, J, Zhang, Z. | | 登録日 | 2021-03-01 | | 公開日 | 2021-06-09 | | 最終更新日 | 2021-07-14 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7E8F
 
 | | SARS-CoV-2 NTD in complex with N9 Fab | | 分子名称: | 368-2 H, 368-2 L, 604 H, ... | | 著者 | Du, S, Xiao, J, Zhang, Z. | | 登録日 | 2021-03-01 | | 公開日 | 2021-06-09 | | 最終更新日 | 2021-07-14 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7E86
 
 | |
7E88
 
 | |
7U3Z
 
 | | [F244] Self-assembling tensegrity triangle with two turns, four turns and four turns of DNA per axis by extension with P1 symmetry | | 分子名称: | DNA (35-MER), DNA (42-MER), DNA (5'-D(*AP*AP*CP*CP*TP*AP*CP*CP*TP*GP*GP*CP*AP*GP*GP*AP*CP*GP*AP*CP*T)-3'), ... | | 著者 | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (7.55 Å) | | 主引用文献 | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U3V
 
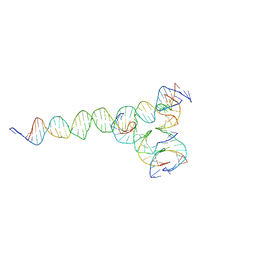 | | [F224] Self-assembling tensegrity triangle with two turns, two turns and four turns of DNA per axis by extension with P1 symmetry | | 分子名称: | DNA (35-MER), DNA (42-MER), DNA (5'-D(*CP*AP*CP*GP*AP*GP*CP*CP*TP*GP*AP*TP*CP*GP*GP*AP*CP*AP*AP*GP*A)-3'), ... | | 著者 | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (5.14 Å) | | 主引用文献 | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U41
 
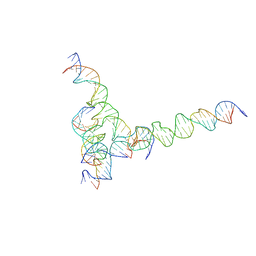 | | [F234] Self-assembling tensegrity triangle with two turns, three turns and four turns of DNA per axis by extension with P1 symmetry | | 分子名称: | DNA (31-MER), DNA (35-MER), DNA (42-MER), ... | | 著者 | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (7.24 Å) | | 主引用文献 | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U42
 
 | | [F334] Self-assembling tensegrity triangle with three turns, three turns and four turns of DNA per axis by extension with P1 symmetry | | 分子名称: | DNA (31-MER), DNA (35-MER), DNA (42-MER), ... | | 著者 | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (7.71 Å) | | 主引用文献 | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U43
 
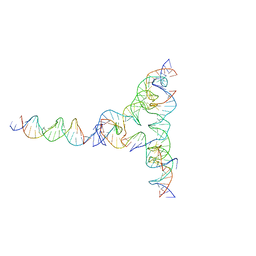 | | [L334] Self-assembling tensegrity triangle with three turns, three turns and four turns of DNA per axis by extension and linker addition with P1 symmetry | | 分子名称: | DNA (31-MER), DNA (5'-D(*TP*AP*CP*AP*CP*CP*GP*AP*TP*CP*AP*CP*CP*TP*GP*CP*CP*AP*CP*CP*G)-3'), DNA (5'-D(P*AP*CP*TP*GP*AP*TP*GP*TP*GP*GP*TP*AP*GP*G)-3'), ... | | 著者 | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (7.55 Å) | | 主引用文献 | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U45
 
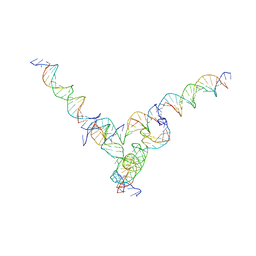 | | [L344] Self-assembling tensegrity triangle with three turns, four turns and four turns of DNA per axis by extension and linker addition with P1 symmetry | | 分子名称: | DNA (31-MER), DNA (5'-D(P*AP*CP*TP*GP*AP*TP*GP*TP*GP*GP*TP*AP*GP*G)-3'), DNA (5'-D(P*AP*GP*GP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), ... | | 著者 | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (8.05 Å) | | 主引用文献 | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U3W
 
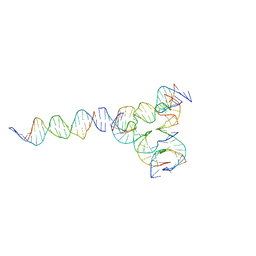 | | [L224] Self-assembling tensegrity triangle with two turns, two turns and four turns of DNA per axis by linker addition with P1 symmetry | | 分子名称: | DNA (5'-D(*AP*AP*CP*CP*TP*AP*CP*CP*TP*GP*GP*CP*AP*GP*GP*AP*CP*GP*AP*CP*T)-3'), DNA (5'-D(*AP*GP*AP*GP*TP*CP*GP*TP*GP*GP*CP*TP*CP*G)-3'), DNA (5'-D(*CP*AP*CP*GP*AP*GP*CP*CP*TP*GP*AP*TP*CP*GP*GP*AP*CP*AP*AP*GP*A)-3'), ... | | 著者 | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (6.33 Å) | | 主引用文献 | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U3S
 
 | | [2T7+21] Self-assembling tensegrity triangle with two turns of DNA and the sticky end addition of a two-turn linker per axis with R3 symmetry | | 分子名称: | DNA (5'-D(P*CP*CP*GP*TP*AP*CP*A)-3'), DNA (5'-D(P*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*GP*AP*TP*GP*CP*TP*GP*AP*CP*GP*TP*AP*GP*TP*AP*GP*CP*AP*GP*AP*G)-3'), ... | | 著者 | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (9.5 Å) | | 主引用文献 | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U3T
 
 | | [F223] Self-assembling tensegrity triangle with two turns, two turns and three turns of DNA per axis by extension with P1 symmetry | | 分子名称: | DNA (31-MER), DNA (5'-D(*AP*AP*CP*CP*TP*AP*CP*CP*TP*GP*GP*CP*AP*GP*GP*AP*CP*GP*AP*CP*T)-3'), DNA (5'-D(*CP*AP*CP*GP*AP*GP*CP*CP*TP*GP*AP*TP*CP*GP*GP*AP*CP*AP*AP*GP*A)-3'), ... | | 著者 | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P, Zhu, E. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (4.69 Å) | | 主引用文献 | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U3O
 
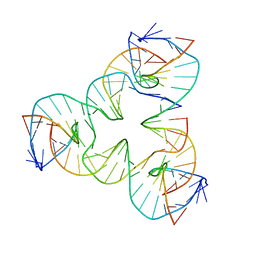 | | [a2T7] Self-assembling asymmetric tensegrity triangle with P1 symmetry | | 分子名称: | DNA (5'-D(*AP*AP*CP*CP*TP*AP*CP*CP*TP*GP*GP*CP*AP*GP*GP*AP*CP*GP*AP*CP*T)-3'), DNA (5'-D(*CP*AP*CP*GP*AP*GP*CP*CP*TP*GP*AP*TP*CP*GP*GP*AP*CP*AP*AP*GP*A)-3'), DNA (5'-D(*GP*AP*GP*CP*GP*AP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), ... | | 著者 | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.31 Å) | | 主引用文献 | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U3U
 
 | | [L223] Self-assembling tensegrity triangle with two turns, two turns and three turns of DNA per axis by linker addition with P1 symmetry | | 分子名称: | DNA (5'-D(*AP*AP*CP*CP*TP*AP*CP*TP*TP*GP*GP*CP*AP*GP*GP*AP*CP*GP*AP*CP*T)-3'), DNA (5'-D(*AP*CP*GP*CP*GP*AP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(*CP*AP*CP*GP*AP*GP*CP*CP*TP*GP*AP*TP*CP*GP*GP*AP*CP*AP*AP*GP*A)-3'), ... | | 著者 | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (4.46 Å) | | 主引用文献 | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
