4EP5
 
 | | Thermus thermophilus RuvC structure | | 分子名称: | Crossover junction endodeoxyribonuclease RuvC, GLYCEROL, SULFATE ION | | 著者 | Chen, L, Shi, K, Yin, Z.Q, Aihara, H. | | 登録日 | 2012-04-17 | | 公開日 | 2012-11-14 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Structural asymmetry in the Thermus thermophilus RuvC dimer suggests a basis for sequential strand cleavages during Holliday junction resolution.
Nucleic Acids Res., 41, 2013
|
|
4FPV
 
 | | Crystal structure of D. rerio TDP2 complexed with single strand DNA product | | 分子名称: | DNA (5'-D(P*TP*GP*CP*AP*G)-3'), GLYCEROL, MAGNESIUM ION, ... | | 著者 | Shi, K, Kurahashi, K, Aihara, H. | | 登録日 | 2012-06-22 | | 公開日 | 2012-10-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Structural basis for recognition of 5'-phosphotyrosine adducts by Tdp2.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4FW2
 
 | |
4FVA
 
 | | Crystal structure of truncated Caenorhabditis elegans TDP2 | | 分子名称: | 1,2-ETHANEDIOL, 5'-tyrosyl-DNA phosphodiesterase, MAGNESIUM ION, ... | | 著者 | Shi, K, Kurahashi, K, Aihara, H. | | 登録日 | 2012-06-29 | | 公開日 | 2012-10-31 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Structural basis for recognition of 5'-phosphotyrosine adducts by Tdp2.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4GEW
 
 | |
4F1I
 
 | |
1UD3
 
 | | Crystal structure of AmyK38 N289H mutant | | 分子名称: | SODIUM ION, amylase | | 著者 | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | 登録日 | 2003-04-28 | | 公開日 | 2003-07-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
1UD8
 
 | | Crystal structure of AmyK38 with lithium ion | | 分子名称: | SODIUM ION, amylase | | 著者 | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | 登録日 | 2003-04-28 | | 公開日 | 2003-07-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.88 Å) | | 主引用文献 | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
1UD2
 
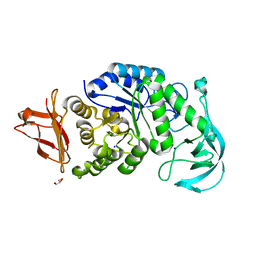 | | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) | | 分子名称: | GLYCEROL, SODIUM ION, amylase | | 著者 | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | 登録日 | 2003-04-28 | | 公開日 | 2003-07-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
3AYX
 
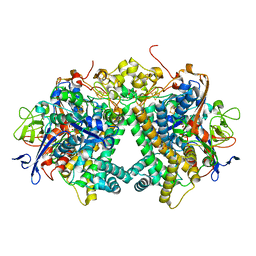 | | Membrane-bound respiratory [NiFe] hydrogenase from Hydrogenovibrio marinus in an H2-reduced condition | | 分子名称: | CARBON MONOXIDE, CYANIDE ION, FE (II) ION, ... | | 著者 | Shomura, Y, Yoon, K.S, Nishihara, H, Higuchi, Y. | | 登録日 | 2011-05-20 | | 公開日 | 2011-10-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.18 Å) | | 主引用文献 | Structural basis for a [4Fe-3S] cluster in the oxygen-tolerant membrane-bound [NiFe]-hydrogenase
Nature, 479, 2011
|
|
3AYZ
 
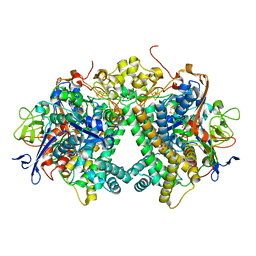 | | Membrane-bound respiratory [NiFe] hydrogenase from Hydrogenovibrio marinus in an air-oxidized condition | | 分子名称: | CARBON MONOXIDE, CYANIDE ION, FE (II) ION, ... | | 著者 | Shomura, Y, Yoon, K.S, Nishihara, H, Higuchi, Y. | | 登録日 | 2011-05-20 | | 公開日 | 2011-10-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | Structural basis for a [4Fe-3S] cluster in the oxygen-tolerant membrane-bound [NiFe]-hydrogenase
Nature, 479, 2011
|
|
1UD6
 
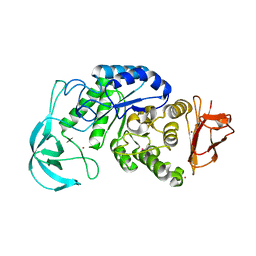 | | Crystal structure of AmyK38 with potassium ion | | 分子名称: | POTASSIUM ION, amylase | | 著者 | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | 登録日 | 2003-04-28 | | 公開日 | 2003-07-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
1UD5
 
 | | Crystal structure of AmyK38 with rubidium ion | | 分子名称: | RUBIDIUM ION, SODIUM ION, amylase | | 著者 | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | 登録日 | 2003-04-28 | | 公開日 | 2003-07-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
1UD4
 
 | | Crystal structure of calcium free alpha amylase from Bacillus sp. strain KSM-K38 (AmyK38, in calcium containing solution) | | 分子名称: | SODIUM ION, amylase | | 著者 | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | 登録日 | 2003-04-28 | | 公開日 | 2003-07-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
2DIE
 
 | | Alkaline alpha-amylase AmyK from Bacillus sp. KSM-1378 | | 分子名称: | CALCIUM ION, SODIUM ION, amylase | | 著者 | Shirai, T, Igarashi, K, Ozawa, T, Hagihara, H, Kobayashi, T, Ozaki, K, Ito, S. | | 登録日 | 2006-03-29 | | 公開日 | 2007-02-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Ancestral sequence evolutionary trace and crystal structure analyses of alkaline alpha-amylase from Bacillus sp. KSM-1378 to clarify the alkaline adaptation process of proteins
Proteins, 66, 2007
|
|
3SIX
 
 | | Crystal structure of NodZ alpha-1,6-fucosyltransferase soaked with GDP-fucose | | 分子名称: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, Nodulation fucosyltransferase NodZ, ... | | 著者 | Brzezinski, K, Dauter, Z, Jaskolski, M. | | 登録日 | 2011-06-20 | | 公開日 | 2012-02-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structures of NodZ alpha-1,6-fucosyltransferase in complex with GDP and GDP-fucose
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3SIW
 
 | | Crystal structure of NodZ alpha-1,6-fucosyltransferase co-crystallized with GDP | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, Nodulation fucosyltransferase NodZ, PHOSPHATE ION | | 著者 | Brzezinski, K, Dauter, Z, Jaskolski, M. | | 登録日 | 2011-06-20 | | 公開日 | 2012-02-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Structures of NodZ alpha-1,6-fucosyltransferase in complex with GDP and GDP-fucose
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3WU4
 
 | | Oxidized-form structure of E.coli Lon Proteolytic domain | | 分子名称: | Lon protease, SULFATE ION | | 著者 | Nishii, W, Kukimoto-Niino, M, Terada, T, Shirouzu, M, Muramatsu, T, Yokoyama, S. | | 登録日 | 2014-04-22 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | A redox switch shapes the Lon protease exit pore to facultatively regulate proteolysis.
Nat. Chem. Biol., 11, 2015
|
|
3A9Z
 
 | |
3A9Y
 
 | |
7M2E
 
 | | Crystal structure of BPTF bromodomain in complex with CB02-092 | | 分子名称: | 4-chloro-5-{4-[2-(dimethylamino)ethyl]anilino}-2-methylpyridazin-3(2H)-one, Nucleosome-remodeling factor subunit BPTF | | 著者 | Nithianantham, S, Fischer, M. | | 登録日 | 2021-03-16 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition
J.Med.Chem., 64, 2021
|
|
7LP0
 
 | |
7LRK
 
 | |
7LRO
 
 | |
7LPK
 
 | |
