5JFP
 
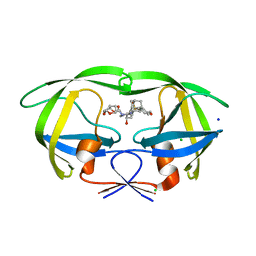 | | HIV-1 wild Type protease with GRL-097-13A (a Adamantane P1-Ligand with bis-THF in P2 and isobutylamine in P1') | | 分子名称: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-[(3R,5R,7R)-tricyclo[3.3.1.1~3,7~]decan-1-yl]butan-2-yl}carbamate, CHLORIDE ION, Protease, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2016-04-19 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Probing Lipophilic Adamantyl Group as the P1-Ligand for HIV-1 Protease Inhibitors: Design, Synthesis, Protein X-ray Structural Studies, and Biological Evaluation.
J.Med.Chem., 59, 2016
|
|
5JFU
 
 | | HIV-1 wild Type protease with GRL-007-14A (a Adamantane P1-Ligand with bis-THF in P2 and benzylamine in P1') | | 分子名称: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-4-{benzyl[(4-methoxyphenyl)sulfonyl]amino}-3-hydroxy-1-[(3R,5R,7R)-tricyclo[3.3.1.1~3,7~]decan-1-yl]butan-2-yl}carbamate, CHLORIDE ION, Protease | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2016-04-19 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Probing Lipophilic Adamantyl Group as the P1-Ligand for HIV-1 Protease Inhibitors: Design, Synthesis, Protein X-ray Structural Studies, and Biological Evaluation.
J.Med.Chem., 59, 2016
|
|
5JG1
 
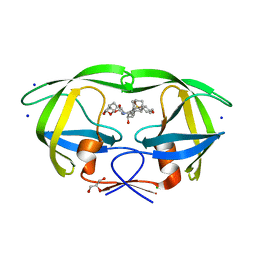 | | HIV-1 wild Type protease with GRL-031-14A (a Adamantane P1-Ligand with tetrahydropyrano-tetrahydrofuran in P2 and isobutylamine in P1') | | 分子名称: | (3R,3aS,7aR)-hexahydro-4H-furo[2,3-b]pyran-3-yl {(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-[(3R,5R,7R)-tricyclo[3.3.1.1~3,7~]decan-1-yl]butan-2-yl}carbamate, CHLORIDE ION, GLYCEROL, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2016-04-19 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Probing Lipophilic Adamantyl Group as the P1-Ligand for HIV-1 Protease Inhibitors: Design, Synthesis, Protein X-ray Structural Studies, and Biological Evaluation.
J.Med.Chem., 59, 2016
|
|
8XO3
 
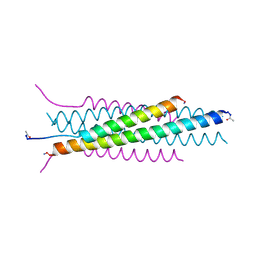 | |
8XO2
 
 | |
8XO7
 
 | |
8XNE
 
 | |
8XO5
 
 | | Crystal structure of measles virus fusion inhibitor MEK28 complexed with F protein HR1 (HR1-40) (H3 space group) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Fusion glycoprotein F1, Measles virus fusion inhibitor MEK28 | | 著者 | Oishi, S, Takahara, A, Nakatsu, T. | | 登録日 | 2023-12-31 | | 公開日 | 2025-01-01 | | 最終更新日 | 2025-03-05 | | 実験手法 | X-RAY DIFFRACTION (1.205 Å) | | 主引用文献 | Elucidation of Postfusion Structures of the Measles Virus F Protein for the Structure-Based Design of Fusion Inhibitors.
J.Med.Chem., 68, 2025
|
|
8XO4
 
 | |
8XO6
 
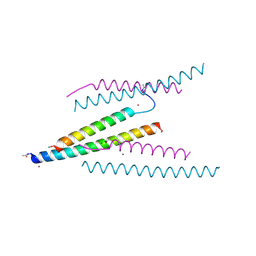 | | Crystal structure of measles virus fusion inhibitor MEK35GE complexed with F protein HR1 (HR1-42) (P21212 space group) | | 分子名称: | ACETATE ION, Fusion glycoprotein F1, Measles virus fusion inhibitor MEK35GE, ... | | 著者 | Oishi, S, Takahara, A, Nakatsu, T. | | 登録日 | 2023-12-31 | | 公開日 | 2025-01-01 | | 最終更新日 | 2025-03-05 | | 実験手法 | X-RAY DIFFRACTION (1.457 Å) | | 主引用文献 | Elucidation of Postfusion Structures of the Measles Virus F Protein for the Structure-Based Design of Fusion Inhibitors.
J.Med.Chem., 68, 2025
|
|
8XO8
 
 | |
5BS4
 
 | | HIV-1 wild Type protease with GRL-047-11A (a methylamine bis-Tetrahydrofuran P2-Ligand, 4-amino sulfonamide derivative) | | 分子名称: | (3R,3aS,4R,6aR)-4-(methylamino)hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2015-06-01 | | 公開日 | 2015-09-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | Design of HIV-1 Protease Inhibitors with Amino-bis-tetrahydrofuran Derivatives as P2-Ligands to Enhance Backbone-Binding Interactions: Synthesis, Biological Evaluation, and Protein-Ligand X-ray Studies.
J.Med.Chem., 58, 2015
|
|
5BRY
 
 | | HIV-1 wild Type protease with GRL-011-11A (a methylamine bis-Tetrahydrofuran P2-Ligand, sulfonamide isostere derivate) | | 分子名称: | (3R,3aS,4R,6aR)-4-(methylamino)hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, Protease, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2015-06-01 | | 公開日 | 2015-09-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | Design of HIV-1 Protease Inhibitors with Amino-bis-tetrahydrofuran Derivatives as P2-Ligands to Enhance Backbone-Binding Interactions: Synthesis, Biological Evaluation, and Protein-Ligand X-ray Studies.
J.Med.Chem., 58, 2015
|
|
1KA9
 
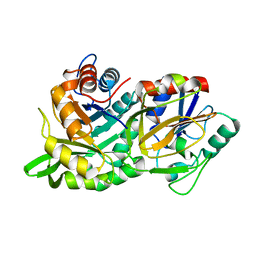 | |
1KZK
 
 | | JE-2147-HIV Protease Complex | | 分子名称: | (4R)-3-{(2S,3S)-2-hydroxy-3-[(3-hydroxy-2-methylbenzoyl)amino]-4-phenylbutanoyl}-5,5-dimethyl-N-(2-methylbenzyl)-1,3-thiazolidine-4-carboxamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Reiling, K.K, Endres, N.F, Dauber, D.S, Craik, C.S, Stroud, R.M. | | 登録日 | 2002-02-06 | | 公開日 | 2002-04-03 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.09 Å) | | 主引用文献 | Anisotropic Dynamics of the JE-2147-HIV Protease Complex:
Drug Resistance and Thermodynamic Binding Mode Examined in a 1.09 A Structure
Biochemistry, 41, 2002
|
|
1IYE
 
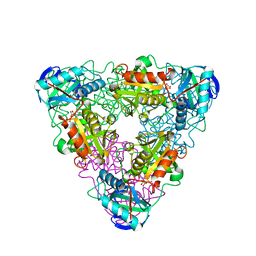 | | CRYSTAL STRUCTURE OF ESCHELICHIA COLI BRANCHED-CHAIN AMINO ACID AMINOTRANSFERASE | | 分子名称: | BRANCHED-CHAIN AMINO ACID AMINOTRANSFERASE, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-glutamic acid | | 著者 | Hirotsu, K, Goto, M. | | 登録日 | 2002-08-07 | | 公開日 | 2003-05-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Crystal structures of branched-chain amino Acid aminotransferase complexed with glutamate and glutarate: true reaction intermediate and double substrate recognition of the enzyme.
Biochemistry, 42, 2003
|
|
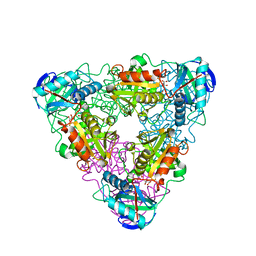
1IYD
 
 | |
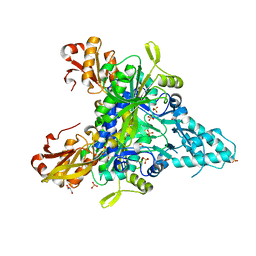
5Z5E
 
 | |
3A9Z
 
 | |
3A2B
 
 | |
3A9Y
 
 | |
2Z5E
 
 | | Crystal Structure of Proteasome Assembling Chaperone 3 | | 分子名称: | Proteasome Assembling Chaperone 3 | | 著者 | Okamoto, K, Kurimoto, E, Sakata, E, Suzuki, A, Yamane, T, Hirano, Y, Murata, S, Tanaka, K, Kato, K. | | 登録日 | 2007-07-06 | | 公開日 | 2008-02-19 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of a chaperone complex that contributes to the assembly of yeast 20S proteasomes
Nat.Struct.Mol.Biol., 15, 2008
|
|
3A9X
 
 | | Crystal structure of rat selenocysteine lyase | | 分子名称: | PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, Selenocysteine lyase | | 著者 | Omi, R, Hirotsu, K. | | 登録日 | 2009-11-08 | | 公開日 | 2010-03-16 | | 最終更新日 | 2025-03-26 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Reaction mechanism and molecular basis for selenium/sulfur discrimination of selenocysteine lyase.
J.Biol.Chem., 285, 2010
|
|
7DJI
 
 | |
7F8K
 
 | |
