1Q87
 
 | |
1RZR
 
 | | crystal structure of transcriptional regulator-phosphoprotein-DNA complex | | 分子名称: | 5'-D(*CP*TP*GP*AP*AP*AP*GP*CP*GP*CP*TP*AP*AP*CP*AP*G)-3', 5'-D(*CP*TP*GP*TP*TP*AP*GP*CP*GP*CP*TP*TP*TP*CP*AP*G)-3', Glucose-resistance amylase regulator, ... | | 著者 | Schumacher, M.A, Allen, G.S, Brennan, R.G. | | 登録日 | 2003-12-27 | | 公開日 | 2004-10-12 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis for allosteric control of the transcription regulator CcpA by the phosphoprotein HPr-Ser46-P.
Cell(Cambridge,Mass.), 118, 2004
|
|
1PP8
 
 | | crystal structure of the T. vaginalis IBP39 Initiator binding domain (IBD) bound to the alpha-SCS Inr element | | 分子名称: | 39 kDa initiator binding protein, ALPHA-SCS INR, SULFATE ION | | 著者 | Schumacher, M.A, Lau, A.O.T, Johnson, P.J. | | 登録日 | 2003-06-16 | | 公開日 | 2003-11-18 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Structural Basis of Core Promoter Recognition in a Primitive Eukaryote
Cell(Cambridge,Mass.), 115, 2003
|
|
1SDL
 
 | | CROSS-LINKED, CARBONMONOXY HEMOGLOBIN A | | 分子名称: | 1,3,5-BENZENETRICARBOXYLIC ACID, CARBON MONOXIDE, HEMOGLOBIN A, ... | | 著者 | Schumacher, M.A, Dixon, M.M, Kluger, R, Jones, R.T, Brennan, R.G. | | 登録日 | 1996-02-26 | | 公開日 | 1996-08-01 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Allosteric transition intermediates modelled by crosslinked haemoglobins.
Nature, 375, 1995
|
|
1SDK
 
 | | CROSS-LINKED, CARBONMONOXY HEMOGLOBIN A | | 分子名称: | 1,3,5-BENZENETRICARBOXYLIC ACID, CARBON MONOXIDE, HEMOGLOBIN A, ... | | 著者 | Schumacher, M.A, Dixon, M.M, Kluger, R, Jones, R.T, Brennan, R.G. | | 登録日 | 1996-02-26 | | 公開日 | 1996-08-01 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Allosteric transition intermediates modelled by crosslinked haemoglobins.
Nature, 375, 1995
|
|
1Q89
 
 | |
1SXH
 
 | | apo structure of B. megaterium transcription regulator | | 分子名称: | Glucose-resistance amylase regulator | | 著者 | Schumacher, M.A, Allen, G.S, Diel, M, Seidel, G, Hillen, W, Brennan, R.G. | | 登録日 | 2004-03-30 | | 公開日 | 2004-10-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural studies on the apo transcription factor form B. megaterium
Cell(Cambridge,Mass.), 118, 2004
|
|
1PP7
 
 | | Crystal structure of the T. vaginalis Initiator binding protein bound to the ferredoxin Inr | | 分子名称: | 39 kDa initiator binding protein, FERREDOXIN INR, ZINC ION | | 著者 | Schumacher, M.A, Lau, A.O.T, Johnson, P.J. | | 登録日 | 2003-06-16 | | 公開日 | 2003-11-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structural Basis of Core Promoter Recognition in a Primitive Eukaryote
Cell(Cambridge,Mass.), 115, 2003
|
|
1SXI
 
 | | Structure of apo transcription regulator B. megaterium | | 分子名称: | Glucose-resistance amylase regulator, MAGNESIUM ION | | 著者 | Schumacher, M.A, Allen, G.S, Diel, M, Seidel, G, Hillen, W, Brennan, R.G. | | 登録日 | 2004-03-30 | | 公開日 | 2004-10-19 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural studies on the apo transcription factor form B. megaterium
Cell(Cambridge,Mass.), 118, 2004
|
|
1SXG
 
 | | Structural studies on the apo transcription factor form B. megaterium | | 分子名称: | 2-PHENYLAMINO-ETHANESULFONIC ACID, Glucose-resistance amylase regulator | | 著者 | Schumacher, M.A, Allen, G.S, Diel, M, Seidel, G, Hillen, W, Brennan, R.G. | | 登録日 | 2004-03-30 | | 公開日 | 2004-10-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural studies on the apo transcription factor form B. megaterium
Cell(Cambridge,Mass.), 118, 2004
|
|
7RMW
 
 | | Crystal structure of B. subtilis PurR bound to ppGpp | | 分子名称: | GUANOSINE-5',3'-TETRAPHOSPHATE, Pur operon repressor | | 著者 | Schumacher, M.A. | | 登録日 | 2021-07-28 | | 公開日 | 2021-12-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | The nucleotide messenger (p)ppGpp is an anti-inducer of the purine synthesis transcription regulator PurR in Bacillus.
Nucleic Acids Res., 50, 2022
|
|
6E4N
 
 | |
6E4O
 
 | |
6E4P
 
 | |
5HT1
 
 | |
5HUA
 
 | |
5HTG
 
 | |
5I98
 
 | |
8DPK
 
 | |
8CSH
 
 | |
4YJ1
 
 | |
6UEP
 
 | |
6UEO
 
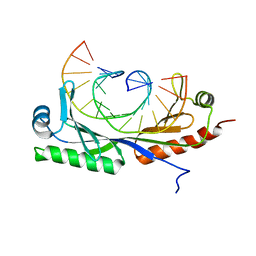 | | Structure of A. thaliana TBP-AC mismatch DNA site | | 分子名称: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*AP*AP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*CP*TP*TP*TP*AP*TP*AP*GP*C)-3'), TATA-box-binding protein 1 | | 著者 | Schumacher, M.A. | | 登録日 | 2019-09-22 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | DNA mismatches reveal conformational penalties in protein-DNA recognition.
Nature, 587, 2020
|
|
6UER
 
 | |
6UEQ
 
 | |
