1P5H
 
 | |
1DAD
 
 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DETHIOBIOTIN SYNTHETASE | | 著者 | Huang, W, Jia, J, Schneider, G, Lindqvist, Y. | | 登録日 | 1995-05-08 | | 公開日 | 1996-06-20 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Mechanism of an ATP-dependent carboxylase, dethiobiotin synthetase, based on crystallographic studies of complexes with substrates and a reaction intermediate.
Biochemistry, 34, 1995
|
|
1DAG
 
 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH 7-(CARBOXYAMINO)-8-AMINO-NONANOIC ACID AND 5'-ADENOSYL-METHYLENE-TRIPHOSPHATE | | 分子名称: | 7-(CARBOXYAMINO)-8-AMINO-NONANOIC ACID, DETHIOBIOTIN SYNTHETASE, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | 著者 | Huang, W, Jia, J, Schneider, G, Lindqvist, Y. | | 登録日 | 1995-05-08 | | 公開日 | 1996-06-20 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Mechanism of an ATP-dependent carboxylase, dethiobiotin synthetase, based on crystallographic studies of complexes with substrates and a reaction intermediate.
Biochemistry, 34, 1995
|
|
1T4C
 
 | | Formyl-CoA Transferase in complex with Oxalyl-CoA | | 分子名称: | COENZYME A, Formyl-CoA:oxalate CoA-transferase, OXALIC ACID | | 著者 | Ricagno, S, Jonsson, S, Richards, N.G, Lindqvist, Y. | | 登録日 | 2004-04-29 | | 公開日 | 2004-08-03 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Kinetic and mechanistic characterization of the formyl-CoA transferase from Oxalobacter formigenes
J.Biol.Chem., 279, 2004
|
|
1STD
 
 | |
1T3Z
 
 | | Formyl-CoA Tranferase mutant Asp169 to Ser | | 分子名称: | Formyl-coenzyme A transferase, OXIDIZED COENZYME A | | 著者 | Ricagno, S, Jonsson, S, Richards, N.G, Lindqvist, Y. | | 登録日 | 2004-04-28 | | 公開日 | 2004-08-03 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Kinetic and mechanistic characterization of the formyl-CoA transferase from Oxalobacter formigenes
J.Biol.Chem., 279, 2004
|
|
7STD
 
 | | SCYTALONE DEHYDRATASE PLUS INHIBITOR 4 | | 分子名称: | ((1RS,3SR)-2,2-DICHLORO-N-[(R)-1-(4-CHLOROPHENYL)ETHYL]-1-ETHYL-3-METHYLCYCLOPROPANECARBOXAMIDE, CALCIUM ION, Scytalone dehydratase | | 著者 | Wawrzak, Z, Sandalova, T, Steffens, J.J, Basarab, G.S, Lundqvist, T, Lindqvist, Y, Jordan, D.B. | | 登録日 | 1999-02-11 | | 公開日 | 1999-12-29 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | High-resolution structures of scytalone dehydratase-inhibitor complexes crystallized at physiological pH.
Proteins, 35, 1999
|
|
2XZ0
 
 | | The Structure of the 2:1 (Partially Occupied) Complex Between Stearoyl Acyl Carrier Protein Desaturase from Ricinus Communis (Castor Bean) and Acyl Carrier Protein. | | 分子名称: | 1,2-ETHANEDIOL, ACYL CARRIER PROTEIN 1, CHLOROPLASTIC, ... | | 著者 | Moche, M, Guy, J.E, Whittle, E, Lengqvist, J, Shanklin, J, Lindqvist, Y. | | 登録日 | 2010-11-22 | | 公開日 | 2011-09-07 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Remote Control of Regioselectivity in Acyl-Acyl Carrier Protein-Desaturases.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1H7X
 
 | | Dihydropyrimidine dehydrogenase (DPD) from pig, ternary complex of a mutant enzyme (C671A), NADPH and 5-fluorouracil | | 分子名称: | 5-FLUOROURACIL, DIHYDROPYRIMIDINE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, ... | | 著者 | Dobritzsch, D, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | 登録日 | 2001-01-19 | | 公開日 | 2001-02-23 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Crystal Structure of Dihydropyrimidine Dehydrogenase, a Major Determinant of the Pharmacokinetics of the Anti-Cancer Drug 5-Fluorouracil
Embo J., 20, 2001
|
|
2CND
 
 | | STRUCTURAL STUDIES ON CORN NITRATE REDUCTASE: REFINED STRUCTURE OF THE CYTOCHROME B REDUCTASE FRAGMENT AT 2.5 ANGSTROMS, ITS ADP COMPLEX AND AN ACTIVE SITE MUTANT AND MODELING OF THE CYTOCHROME B DOMAIN | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-DEPENDENT NITRATE REDUCTASE | | 著者 | Lu, G, Lindqvist, Y, Schneider, G. | | 登録日 | 1995-02-01 | | 公開日 | 1995-04-20 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural studies on corn nitrate reductase: refined structure of the cytochrome b reductase fragment at 2.5 A, its ADP complex and an active-site mutant and modeling of the cytochrome b domain.
J.Mol.Biol., 248, 1995
|
|
5OLT
 
 | | Crystal structure of the extramembrane domain of the cellulose biosynthetic protein BcsG from Salmonella typhimurium | | 分子名称: | CITRIC ACID, Cellulose biosynthesis protein BcsG, ZINC ION | | 著者 | Vella, P, Polyakova, A, Lindqvist, Y, Schnell, R, Bourenkov, G, Schneider, T, Schneider, G. | | 登録日 | 2017-07-28 | | 公開日 | 2018-08-08 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structural and Functional Characterization of the BcsG Subunit of the Cellulose Synthase in Salmonella typhimurium.
J. Mol. Biol., 430, 2018
|
|
1GT8
 
 | | DIHYDROPYRIMIDINE DEHYDROGENASE (DPD) FROM PIG, TERNARY COMPLEX WITH NADPH AND URACIL-4-ACETIC ACID | | 分子名称: | DIHYDROPYRIMIDINE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Dobritzsch, D, Ricagno, S, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | 登録日 | 2002-01-14 | | 公開日 | 2002-04-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.
J. Biol. Chem., 277, 2002
|
|
1GTE
 
 | | DIHYDROPYRIMIDINE DEHYDROGENASE (DPD) FROM PIG, BINARY COMPLEX WITH 5-IODOURACIL | | 分子名称: | 5-IODOURACIL, DIHYDROPYRIMIDINE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, ... | | 著者 | Dobritzsch, D, Ricagno, S, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | 登録日 | 2002-01-15 | | 公開日 | 2002-04-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.
J. Biol. Chem., 277, 2002
|
|
1GTH
 
 | | DIHYDROPYRIMIDINE DEHYDROGENASE (DPD) FROM PIG, TERNARY COMPLEX WITH NADPH AND 5-IODOURACIL | | 分子名称: | (5S)-5-IODODIHYDRO-2,4(1H,3H)-PYRIMIDINEDIONE, 5-IODOURACIL, DIHYDROPYRIMIDINE DEHYDROGENASE, ... | | 著者 | Dobritzsch, D, Ricagno, S, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | 登録日 | 2002-01-15 | | 公開日 | 2002-04-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.
J. Biol. Chem., 277, 2002
|
|
2XZ1
 
 | | The Structure of the 2:2 (Fully Occupied) Complex Between Stearoyl Acyl Carrier Protein Desaturase from Ricinus Communis (Castor Bean) and Acyl Carrier Protein. | | 分子名称: | (2R)-2,4-dihydroxy-3,3-dimethyl-N-{3-oxo-3-[(2-sulfanylethyl)amino]propyl}butanamide, ACYL CARRIER PROTEIN 1, CHLOROPLASTIC, ... | | 著者 | Guy, J.E, Moche, M, Whittle, E, Lengqvist, J, Shanklin, J, Lindqvist, Y. | | 登録日 | 2010-11-22 | | 公開日 | 2011-09-07 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Remote Control of Regioselectivity in Acyl-Acyl Carrier Protein-Desaturases.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1E5L
 
 | | Apo saccharopine reductase from Magnaporthe grisea | | 分子名称: | SACCHAROPINE REDUCTASE | | 著者 | Johansson, E, Steffens, J.J, Lindqvist, Y, Schneider, G. | | 登録日 | 2000-07-27 | | 公開日 | 2000-11-27 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure of Saccharopine Reductase from Magnaporthe Grisea, an Enzyme of the Alpha-Aminoadipate Pathway of Lysine Biosynthesis
Structure, 8, 2000
|
|
5OJH
 
 | | Crystal structure of the extramembrane domain of the cellulose biosynthetic protein BcsG from Salmonella typhimurium | | 分子名称: | CITRATE ANION, Cellulose biosynthesis protein BcsG, ZINC ION | | 著者 | Schneider, G, Vella, P, Lindqvist, Y, Schnell, R. | | 登録日 | 2017-07-21 | | 公開日 | 2018-08-08 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural and Functional Characterization of the BcsG Subunit of the Cellulose Synthase in Salmonella typhimurium.
J. Mol. Biol., 430, 2018
|
|
1FF9
 
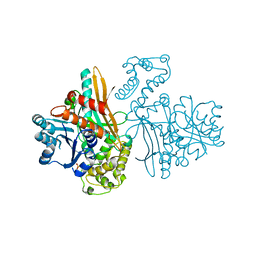 | | APO SACCHAROPINE REDUCTASE | | 分子名称: | SACCHAROPINE REDUCTASE, SULFATE ION | | 著者 | Johansson, E, Steffens, J.J, Lindqvist, Y, Schneider, G. | | 登録日 | 2000-07-25 | | 公開日 | 2000-11-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of saccharopine reductase from Magnaporthe grisea, an enzyme of the alpha-aminoadipate pathway of lysine biosynthesis.
Structure Fold.Des., 8, 2000
|
|
1FOH
 
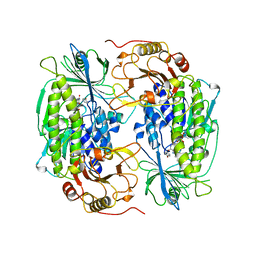 | | PHENOL HYDROXYLASE FROM TRICHOSPORON CUTANEUM | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, PHENOL, PHENOL HYDROXYLASE | | 著者 | Enroth, C, Neujahr, H, Schneider, G, Lindqvist, Y. | | 登録日 | 1998-03-26 | | 公開日 | 1998-06-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The crystal structure of phenol hydroxylase in complex with FAD and phenol provides evidence for a concerted conformational change in the enzyme and its cofactor during catalysis.
Structure, 6, 1998
|
|
1DTS
 
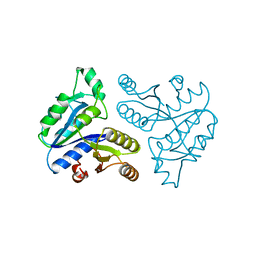 | | CRYSTAL STRUCTURE OF AN ATP DEPENDENT CARBOXYLASE, DETHIOBIOTIN SYNTHASE, AT 1.65 ANGSTROMS RESOLUTION | | 分子名称: | DETHIOBIOTIN SYNTHETASE | | 著者 | Huang, W, Lindqvist, Y, Schneider, G. | | 登録日 | 1995-03-28 | | 公開日 | 1995-04-20 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of an ATP-dependent carboxylase, dethiobiotin synthetase, at 1.65 A resolution.
Structure, 2, 1994
|
|
2VJM
 
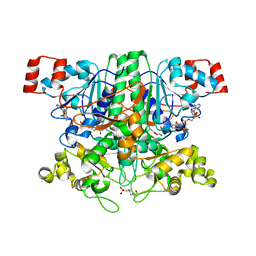 | | Formyl-CoA transferase with aspartyl-formyl anhydide intermediate | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, COENZYME A, FORMYL-COENZYME A TRANSFERASE, ... | | 著者 | Berthold, C.L, Toyota, C.G, Richards, N.G.J, Lindqvist, Y. | | 登録日 | 2007-12-11 | | 公開日 | 2007-12-25 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Reinvestigation of the Catalytic Mechanism of Formyl-Coa Transferase, a Class III Coa-Transferase.
J.Biol.Chem., 283, 2008
|
|
2VJK
 
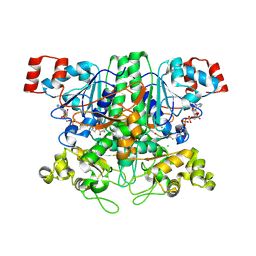 | | Formyl-CoA transferase with aspartyl-CoA thioester intermediate derived from oxalyl-CoA | | 分子名称: | CHLORIDE ION, COENZYME A, FORMYL-COENZYME A TRANSFERASE, ... | | 著者 | Berthold, C.L, Toyota, C.G, Richards, N.G.J, Lindqvist, Y. | | 登録日 | 2007-12-11 | | 公開日 | 2007-12-25 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Reinvestigation of the Catalytic Mechanism of Formyl-Coa Transferase, a Class III Coa-Transferase.
J.Biol.Chem., 283, 2008
|
|
2VJP
 
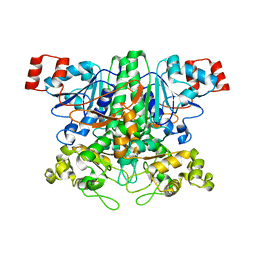 | | Formyl-CoA transferase mutant variant W48F | | 分子名称: | FORMYL-COENZYME A TRANSFERASE, SODIUM ION | | 著者 | Toyota, C.G, Berthold, C.L, Gruez, A, Jonsson, S, Lindqvist, Y, Cambillau, C, Richards, N.G.J. | | 登録日 | 2007-12-11 | | 公開日 | 2008-01-15 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Differential Substrate Specificity and Kinetic Behavior of Escherichia Coli Yfdw and Oxalobacter Formigenes Formyl Coenzyme a Transferase.
J.Bacteriol., 190, 2008
|
|
2VRG
 
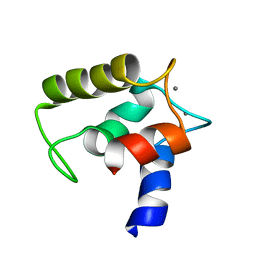 | | Structure of human MCFD2 | | 分子名称: | CALCIUM ION, MULTIPLE COAGULATION FACTOR DEFICIENCY PROTEIN 2 | | 著者 | Guy, J.E, Wigren, E, Svard, M, Hard, T, Lindqvist, Y. | | 登録日 | 2008-04-04 | | 公開日 | 2008-07-15 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | New insights into multiple coagulation factor deficiency from the solution structure of human MCFD2.
J. Mol. Biol., 381, 2008
|
|
1H6V
 
 | | Mammalian thioredoxin reductase | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, THIOREDOXIN REDUCTASE | | 著者 | Sandalova, T, Zhong, L, Lindqvist, Y, Holmgren, A, Schneider, G. | | 登録日 | 2001-06-27 | | 公開日 | 2001-08-14 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Three-Dimensional Structure of a Mammalian Thioredoxin Reductase: Implication for Mechanism and Evolution of a Selenocysteine Dependent Enzyme
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
