7MP6
 
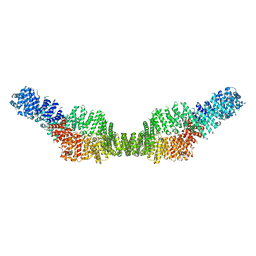 | | Neurofibromin homodimer | | 分子名称: | Isoform I of Neurofibromin | | 著者 | Lupton, C.J, Bayly-Jones, C, Ellisdon, A.M. | | 登録日 | 2021-05-04 | | 公開日 | 2021-12-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (6.25 Å) | | 主引用文献 | The cryo-EM structure of the human neurofibromin dimer reveals the molecular basis for neurofibromatosis type 1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MOC
 
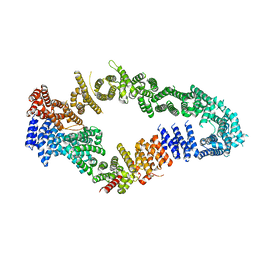 | | Neurofibromin core | | 分子名称: | Isoform I of Neurofibromin | | 著者 | Lupton, C.J, Bayly-Jones, C, Ellisdon, A.M. | | 登録日 | 2021-05-01 | | 公開日 | 2021-12-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (4.56 Å) | | 主引用文献 | The cryo-EM structure of the human neurofibromin dimer reveals the molecular basis for neurofibromatosis type 1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
9ISH
 
 | |
7QZ8
 
 | |
7QZ7
 
 | |
7QZ6
 
 | |
7QZ9
 
 | |
7QZ5
 
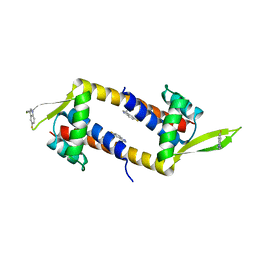 | |
8WTI
 
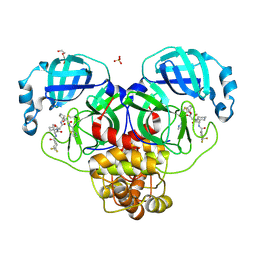 | | Crystal structure of the SARS-CoV-2 main protease in complex with 20j | | 分子名称: | 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | 著者 | Zeng, R, Zhao, X, Yang, S.Y, Lei, J. | | 登録日 | 2023-10-18 | | 公開日 | 2024-08-28 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Discovery of alpha-Ketoamide inhibitors of SARS-CoV-2 main protease derived from quaternized P1 groups.
Bioorg.Chem., 143, 2024
|
|
1YT5
 
 | |
2H4N
 
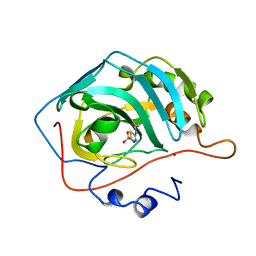 | | H94N CARBONIC ANHYDRASE II COMPLEXED WITH ACETAZOLAMIDE | | 分子名称: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CARBONIC ANHYDRASE II, ZINC ION | | 著者 | Lesburg, C.A, Christianson, D.W. | | 登録日 | 1997-05-29 | | 公開日 | 1997-09-17 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Histidine --> carboxamide ligand substitutions in the zinc binding site of carbonic anhydrase II alter metal coordination geometry but retain catalytic activity.
Biochemistry, 36, 1997
|
|
2I6W
 
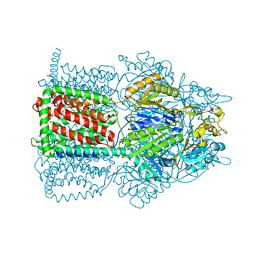 | |
1HTV
 
 | |
1T6S
 
 | | Crystal structure of a conserved hypothetical protein from Chlorobium tepidum | | 分子名称: | NITRATE ION, conserved hypothetical protein | | 著者 | Kim, J.S, Shin, D.H, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | 登録日 | 2004-05-07 | | 公開日 | 2004-12-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of ScpB from Chlorobium tepidum, a protein involved in chromosome partitioning.
Proteins, 62, 2006
|
|
7DGW
 
 | | De novo designed protein H4A2S | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, de novo designed protein H4A2S | | 著者 | Xu, Y, Liao, S, Chen, Q, Liu, H. | | 登録日 | 2020-11-12 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7DGU
 
 | |
7DKK
 
 | | De novo design protein XM2H | | 分子名称: | De novo design protein XM2H | | 著者 | Bin, H. | | 登録日 | 2020-11-24 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7DKO
 
 | | De novo design protein AM2M | | 分子名称: | de novo designed protein AM2M | | 著者 | Bin, H. | | 登録日 | 2020-11-25 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7DMF
 
 | |
6NII
 
 | |
7DGY
 
 | |
6NJD
 
 | |
7CI3
 
 | |
7FBD
 
 | | De novo design protein D53 with MBP tag | | 分子名称: | Maltodextrin-binding protein,De novo design protein D53 | | 著者 | Bin, H. | | 登録日 | 2021-07-09 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7FBB
 
 | | De novo design protein D12 with MBP tag | | 分子名称: | Maltodextrin-binding protein,de novo designed protein D12 | | 著者 | Bin, H. | | 登録日 | 2021-07-09 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.307 Å) | | 主引用文献 | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
