8GA9
 
 | |
8G8I
 
 | |
8GAQ
 
 | |
8GAA
 
 | |
8GEL
 
 | | Cryo-EM structure of synthetic tetrameric building block sC4 | | 分子名称: | sC4 | | 著者 | Redler, R.L, Huddy, T.F, Hsia, Y, Baker, D, Ekiert, D, Bhabha, G. | | 登録日 | 2023-03-07 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Blueprinting extendable nanomaterials with standardized protein blocks.
Nature, 627, 2024
|
|
8GLT
 
 | | Backbone model of de novo-designed chlorophyll-binding nanocage O32-15 | | 分子名称: | C2-chlorophyll-comp_O32-15_ctermHis, polyalanine model, C3-comp_O32-15 | | 著者 | Redler, R.L, Ennist, N.M, Wang, S, Baker, D, Ekiert, D.C, Bhabha, G. | | 登録日 | 2023-03-23 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | De novo design of proteins housing excitonically coupled chlorophyll special pairs.
Nat.Chem.Biol., 20, 2024
|
|
8TCF
 
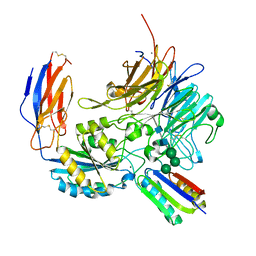 | | Integrin alpha-v beta-8 in complex with minibinder B8_BP_dsulf | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Campbell, M.G, Fernandez, A, Roy, A, Kraft, J, Baker, D. | | 登録日 | 2023-06-30 | | 公開日 | 2023-09-27 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | De novo design of highly selective miniprotein inhibitors of integrins alpha v beta 6 and alpha v beta 8.
Nat Commun, 14, 2023
|
|
8TCG
 
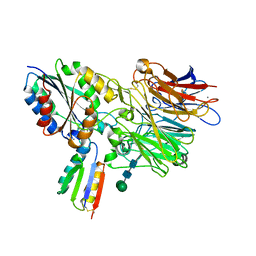 | | Integrin alpha-v beta-6 in complex with minibinder B6_BP_dslf | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Integrin alpha-V heavy chain, ... | | 著者 | Campbell, M.G, Fernandez, A, Roy, A, Kraft, J, Baker, D. | | 登録日 | 2023-06-30 | | 公開日 | 2023-09-27 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | De novo design of highly selective miniprotein inhibitors of integrins alpha v beta 6 and alpha v beta 8.
Nat Commun, 14, 2023
|
|
8SZZ
 
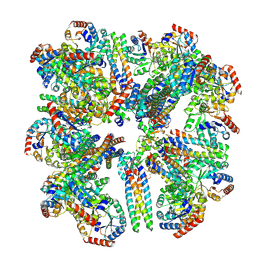 | |
8TL7
 
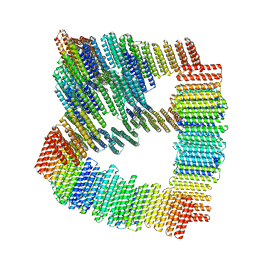 | |
6WSJ
 
 | |
6X1K
 
 | | Solution NMR structure of de novo designed TMB2.3 | | 分子名称: | De novo designed transmembrane beta-barrel TMB2.3 | | 著者 | Liang, B, Vorobieva, A.A, Chow, C.M, Baker, D, Tamm, L.K. | | 登録日 | 2020-05-19 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | De novo design of transmembrane beta barrels.
Science, 371, 2021
|
|
6XNS
 
 | | C3_crown-05 | | 分子名称: | C3_crown-05 | | 著者 | Bick, M.J, Hsia, Y, Sankaran, B, Baker, D. | | 登録日 | 2020-07-04 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.19 Å) | | 主引用文献 | Design of multi-scale protein complexes by hierarchical building block fusion.
Nat Commun, 12, 2021
|
|
6XT4
 
 | |
6XSS
 
 | | CryoEM structure of designed helical fusion protein C4_nat_HFuse-7900 | | 分子名称: | C4_nat_HFuse-7900 | | 著者 | Redler, R.L, Edman, N.I, Baker, D, Ekiert, D, Bhabha, G. | | 登録日 | 2020-07-16 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Design of multi-scale protein complexes by hierarchical building block fusion.
Nat Commun, 12, 2021
|
|
7UBF
 
 | |
7UBD
 
 | |
7UBI
 
 | |
7UCP
 
 | | computationally designed macrocycle | | 分子名称: | computationally designed cyclic peptide D8.3.p2 | | 著者 | Bhardwaj, G, Baker, D, Rettie, S, Glynn, C, Sawaya, M. | | 登録日 | 2022-03-17 | | 公開日 | 2022-09-14 | | 最終更新日 | 2022-09-28 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | Accurate de novo design of membrane-traversing macrocycles.
Cell, 185, 2022
|
|
7UZL
 
 | |
7UBC
 
 | |
7UBE
 
 | |
7UBG
 
 | |
7UBH
 
 | |
7UNJ
 
 | | De novo designed chlorophyll dimer protein with Zn pheophorbide a methyl ester matching geometry of purple bacterial special pair, SP1-ZnPPaM | | 分子名称: | 1,2-ETHANEDIOL, SP1-ZnPPaM designed chlorophyll dimer protein, SULFATE ION, ... | | 著者 | Kennedy, M.A, Stoddard, B.L, Ennist, N.M. | | 登録日 | 2022-04-11 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | De novo design of proteins housing excitonically coupled chlorophyll special pairs.
Nat.Chem.Biol., 2024
|
|
