3X2V
 
 | | Michaelis-like complex of cAMP-dependent Protein Kinase Catalytic Subunit | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CARBONATE ION, MAGNESIUM ION, ... | | 著者 | Das, A, Langan, P, Gerlits, O, Kovalevsky, A.Y, Heller, W.T. | | 登録日 | 2015-01-02 | | 公開日 | 2015-12-16 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Protein Kinase A Catalytic Subunit Primed for Action: Time-Lapse Crystallography of Michaelis Complex Formation.
Structure, 23, 2015
|
|
3X2U
 
 | | Michaelis-like initial complex of cAMP-dependent Protein Kinase Catalytic Subunit. | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Substrate Peptide, ... | | 著者 | Das, A, Langan, P, Gerlits, O, Kovalevsky, A.Y, Heller, W.T. | | 登録日 | 2015-01-02 | | 公開日 | 2015-12-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Protein Kinase A Catalytic Subunit Primed for Action: Time-Lapse Crystallography of Michaelis Complex Formation.
Structure, 23, 2015
|
|
6K9R
 
 | |
5E5K
 
 | | Joint X-ray/neutron structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with darunavir at pH 4.3 | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, HIV-1 protease | | 著者 | Kovalevsky, A.Y, Das, A. | | 登録日 | 2015-10-08 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-03-06 | | 実験手法 | NEUTRON DIFFRACTION (1.75 Å), X-RAY DIFFRACTION | | 主引用文献 | Long-Range Electrostatics-Induced Two-Proton Transfer Captured by Neutron Crystallography in an Enzyme Catalytic Site.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
4K9F
 
 | | Neutron structure of Perdeuterated Rubredoxin refined against 1.75 resolution data collected on the new IMAGINE instrument at HFIR, ORNL | | 分子名称: | FE (III) ION, Rubredoxin | | 著者 | Munshi, P, Meilleur, F, Myles, D. | | 登録日 | 2013-04-19 | | 公開日 | 2013-12-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | NEUTRON DIFFRACTION (1.75 Å) | | 主引用文献 | The IMAGINE instrument: first neutron protein structure and new capabilities for neutron macromolecular crystallography.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5CCD
 
 | |
5CCE
 
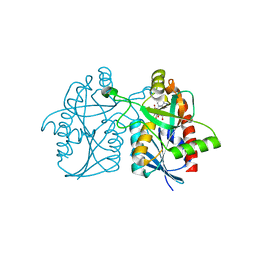 | | Joint X-ray/neutron structure of wild type MTAN complexed with SRH and adenine | | 分子名称: | 5'-Methylthioadenosine Nucleosidase, ADENINE, S-ribosylhomocysteine, ... | | 著者 | Banco, M.T, Kovalevsky, A.Y, Ronning, D.R. | | 登録日 | 2015-07-02 | | 公開日 | 2016-11-16 | | 最終更新日 | 2023-09-27 | | 実験手法 | NEUTRON DIFFRACTION (2.5 Å), X-RAY DIFFRACTION | | 主引用文献 | Neutron structures of the Helicobacter pylori 5'-methylthioadenosine nucleosidase highlight proton sharing and protonation states.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5E5J
 
 | | Joint X-ray/neutron structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with darunavir at pH 6.0 | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | 著者 | Kovalevsky, A.Y, Gerlits, O.O. | | 登録日 | 2015-10-08 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-03-06 | | 実験手法 | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | 主引用文献 | Long-Range Electrostatics-Induced Two-Proton Transfer Captured by Neutron Crystallography in an Enzyme Catalytic Site.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5EBJ
 
 | | Joint X-ray/neutron structure of reversibly photoswitching chromogenic protein, Dathail | | 分子名称: | photoswitching chromogenic protein | | 著者 | Kovalevsky, A.Y, Langan, P.S, Bradbury, A.R.M. | | 登録日 | 2015-10-19 | | 公開日 | 2016-04-06 | | 最終更新日 | 2023-11-15 | | 実験手法 | NEUTRON DIFFRACTION (2.5 Å), X-RAY DIFFRACTION | | 主引用文献 | Evolution and characterization of a new reversibly photoswitching chromogenic protein, Dathail.
J.Mol.Biol., 428, 2016
|
|
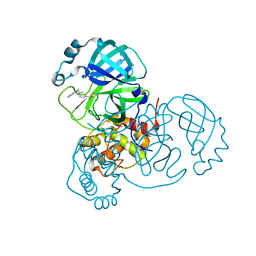
8DL9
 
 | |
8DLB
 
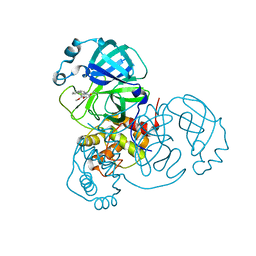 | | Room temperature X-ray structure of SARS-CoV-2 main protease in complex with compound Z2799209083 | | 分子名称: | 1-[(5S)-5-(3,4-dimethoxyphenyl)-3-phenyl-4,5-dihydro-1H-pyrazol-1-yl]ethan-1-one, 3C-like proteinase | | 著者 | Kovalevsky, A.Y, Coates, L, Kneller, D.W. | | 登録日 | 2022-07-07 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | AI-Accelerated Design of Targeted Covalent Inhibitors for SARS-CoV-2.
J.Chem.Inf.Model., 63, 2023
|
|
5ZO0
 
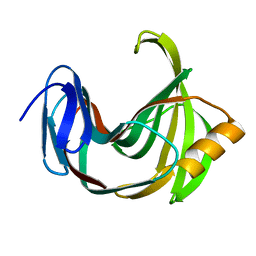 | |
5ZKZ
 
 | |
5ZIW
 
 | |
7LB7
 
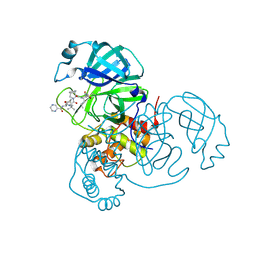 | | Joint X-ray/neutron structure of SARS-CoV-2 main protease (3CL Mpro) in complex with Telaprevir | | 分子名称: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase | | 著者 | Kovalevsky, A.Y, Kneller, D.W, Coates, L. | | 登録日 | 2021-01-07 | | 公開日 | 2021-01-20 | | 最終更新日 | 2024-04-03 | | 実験手法 | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | 主引用文献 | Direct Observation of Protonation State Modulation in SARS-CoV-2 Main Protease upon Inhibitor Binding with Neutron Crystallography.
J.Med.Chem., 64, 2021
|
|
8EYS
 
 | |
8DT4
 
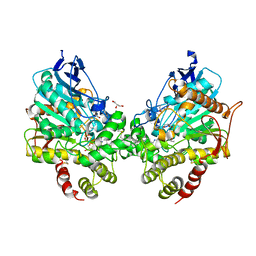 | | X-ray structure of human acetylcholinesterase ternary complex with paraoxon and oxime MMB4 (POX-hAChE-MMB4) | | 分子名称: | 1,1'-methylenebis{4-[(E)-(hydroxyimino)methyl]pyridin-1-ium}, Acetylcholinesterase, DIETHYL PHOSPHONATE, ... | | 著者 | Kovalevsky, A.Y, Gerlits, O, Radic, Z. | | 登録日 | 2022-07-25 | | 公開日 | 2022-11-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural and dynamic effects of paraoxon binding to human acetylcholinesterase by X-ray crystallography and inelastic neutron scattering.
Structure, 30, 2022
|
|
8DT2
 
 | | X-ray structure of human acetylcholinesterase inhibited by paraoxon (POX-hAChE) | | 分子名称: | Acetylcholinesterase, DIETHYL PHOSPHONATE, DIMETHYL SULFOXIDE, ... | | 著者 | Kovalevsky, A.Y, Gerlits, O, Radic, Z. | | 登録日 | 2022-07-25 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.803 Å) | | 主引用文献 | Structural and dynamic effects of paraoxon binding to human acetylcholinesterase by X-ray crystallography and inelastic neutron scattering.
Structure, 30, 2022
|
|
8DT7
 
 | | X-ray structure of human acetylcholinesterase in complex with oxime MMB4 (hAChE-MMB4) | | 分子名称: | 1,1'-methylenebis{4-[(E)-(hydroxyimino)methyl]pyridin-1-ium}, Acetylcholinesterase, GLYCEROL, ... | | 著者 | Kovalevsky, A.Y, Gerlits, O, Radic, Z. | | 登録日 | 2022-07-25 | | 公開日 | 2022-11-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.207 Å) | | 主引用文献 | Structural and dynamic effects of paraoxon binding to human acetylcholinesterase by X-ray crystallography and inelastic neutron scattering.
Structure, 30, 2022
|
|
8DT5
 
 | | X-ray structure of human acetylcholinesterase ternary complex with paraoxon and oxime RS170B (POX-hAChE-RS170B) | | 分子名称: | 4-carbamoyl-1-(3-{2-[(E)-(hydroxyimino)methyl]-1H-imidazol-1-yl}propyl)pyridin-1-ium, Acetylcholinesterase, DIETHYL PHOSPHONATE, ... | | 著者 | Kovalevsky, A.Y, Gerlits, O, Radic, Z. | | 登録日 | 2022-07-25 | | 公開日 | 2022-11-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural and dynamic effects of paraoxon binding to human acetylcholinesterase by X-ray crystallography and inelastic neutron scattering.
Structure, 30, 2022
|
|
5T8H
 
 | |
5VK7
 
 | |
5VJZ
 
 | | Joint X-ray/neutron structure of aspartate aminotransferase with alpha-methyl-aspartate at pH 7.5 | | 分子名称: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-2-METHYL-SUCCINIC ACID, Aspartate aminotransferase, cytoplasmic | | 著者 | Dajnowicz, S, Kovalevsky, A.Y, Mueser, T.C. | | 登録日 | 2017-04-20 | | 公開日 | 2017-11-01 | | 最終更新日 | 2022-03-16 | | 実験手法 | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | 主引用文献 | Direct visualization of critical hydrogen atoms in a pyridoxal 5'-phosphate enzyme.
Nat Commun, 8, 2017
|
|
6JWB
 
 | | Crystal Structures of Endo-beta-1,4-xylanase II Complexed with Xylotriose | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase 2, IODIDE ION, ... | | 著者 | Li, C, Wan, Q. | | 登録日 | 2019-04-19 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Studying the Role of a Single Mutation of a Family 11 Glycoside Hydrolase Using High-Resolution X-ray Crystallography.
Protein J., 39, 2020
|
|
6K9W
 
 | |
