2VTJ
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design | | 分子名称: | 4-[(6-chloropyrazin-2-yl)amino]benzenesulfonamide, CELL DIVISION PROTEIN KINASE 2, GLYCEROL | | 著者 | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | 登録日 | 2008-05-15 | | 公開日 | 2008-08-05 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
2VTT
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design. | | 分子名称: | 4-{[(2,6-difluorophenyl)carbonyl]amino}-N-[(3S)-piperidin-3-yl]-1H-pyrazole-3-carboxamide, CELL DIVISION PROTEIN KINASE 2 | | 著者 | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | 登録日 | 2008-05-15 | | 公開日 | 2008-08-05 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
2VTP
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design. | | 分子名称: | CELL DIVISION PROTEIN KINASE 2, {[(2,6-difluorophenyl)carbonyl]amino}-N-(4-fluorophenyl)-1H-pyrazole-3-carboxamide | | 著者 | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | 登録日 | 2008-05-15 | | 公開日 | 2008-08-05 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
2VTH
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H-pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design | | 分子名称: | 5-hydroxynaphthalene-1-sulfonamide, CELL DIVISION PROTEIN KINASE 2, GLYCEROL | | 著者 | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | 登録日 | 2008-05-15 | | 公開日 | 2008-08-05 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
8C66
 
 | |
2VWY
 
 | | ephB4 kinase domain inhibitor complex | | 分子名称: | EPHRIN TYPE-B RECEPTOR 4, MAGNESIUM ION, N'-(5-chloro-1,3-benzodioxol-4-yl)-N-(3-methylsulfonylphenyl)pyrimidine-2,4-diamine | | 著者 | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L, Barratt, D, Leach, A.G, Kettle, J.G. | | 登録日 | 2008-06-30 | | 公開日 | 2008-10-28 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Inhibitors of the Tyrosine Kinase Ephb4. Part 2: Structure-Based Discovery and Optimisation of 3,5-Bis Substituted Anilinopyrimidines.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2VTM
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design. | | 分子名称: | CELL DIVISION PROTEIN KINASE 2, PYRAZOLO[1,5-A]PYRIMIDINE-3-CARBONITRILE | | 著者 | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | 登録日 | 2008-05-15 | | 公開日 | 2008-08-05 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
2VU3
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design. | | 分子名称: | 4-{[(2,6-dichlorophenyl)carbonyl]amino}-N-piperidin-4-yl-1H-pyrazole-3-carboxamide, CELL DIVISION PROTEIN KINASE 2 | | 著者 | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | 登録日 | 2008-05-20 | | 公開日 | 2008-08-05 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
3NEW
 
 | | p38-alpha complexed with Compound 10 | | 分子名称: | 4-(trifluoromethyl)-3-[3-(trifluoromethyl)phenyl]-1,7-dihydro-6H-pyrazolo[3,4-b]pyridin-6-one, Mitogen-activated protein kinase 14 | | 著者 | Goedken, E.R, Comess, K.M, Sun, C, Argiriadi, M, Jia, Y, Quinn, C.M, Banach, D.L, Marcotte, D, Borhani, D. | | 登録日 | 2010-06-09 | | 公開日 | 2010-12-01 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Discovery and Characterization of Non-ATP Site Inhibitors of the Mitogen Activated Protein (MAP) Kinases.
Acs Chem.Biol., 6, 2011
|
|
2VWT
 
 | | Crystal structure of YfaU, a metal ion dependent class II aldolase from Escherichia coli K12 - Mg-pyruvate product complex | | 分子名称: | GLYCEROL, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Rea, D, Rakus, J.F, Gerlt, J.A, Fulop, V, Bugg, T.D.H, Roper, D.I. | | 登録日 | 2008-06-26 | | 公開日 | 2008-09-09 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Crystal Structure and Functional Assignment of Yfau, a Metal Ion Dependent Class II Aldolase from Escherichia Coli K12.
Biochemistry, 47, 2008
|
|
6NB5
 
 | | Crystal structure of anti- MERS-CoV human neutralizing LCA60 antibody Fab fragment | | 分子名称: | LCA60 antigen-binding (Fab) fragment, heavy chain, light chain | | 著者 | Walls, A.J, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, J, Quispe, J, Cameroni, E, Gopal, R, Dai, M, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2018-12-06 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
7TAS
 
 | |
1K54
 
 | | OXA-10 class D beta-lactamase partially acylated with reacted 6beta-(1-hydroxy-1-methylethyl) penicillanic acid | | 分子名称: | (1R)-2-(1-CARBOXY-2-HYDROXY-2-METHYL-PROPYL)-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, 1,2-ETHANEDIOL, Beta lactamase OXA-10, ... | | 著者 | Golemi, D, Maveyraud, L, Vakulenko, S, Samama, J.P, Mobashery, S. | | 登録日 | 2001-10-10 | | 公開日 | 2001-11-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Critical involvement of a carbamylated lysine in catalytic function of class D beta-lactamases.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
7TAT
 
 | |
6NB8
 
 | | Crystal structure of anti- SARS-CoV human neutralizing S230 antibody Fab fragment | | 分子名称: | S230 antigen-binding (Fab) fragment, heavy chain, light chain | | 著者 | Walls, A.J, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, J, Quispe, J, Cameroni, E, Gopal, R, Dai, M, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2018-12-06 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
1K57
 
 | | OXA 10 class D beta-lactamase at pH 6.0 | | 分子名称: | BETA LACTAMASE OXA-10, SULFATE ION | | 著者 | Golemi, D, Maveyraud, L, Vakulenko, S, Samama, J.P, Mobashery, S. | | 登録日 | 2001-10-10 | | 公開日 | 2001-11-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Critical involvement of a carbamylated lysine in catalytic function of class D beta-lactamases.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1K56
 
 | | OXA 10 class D beta-lactamase at pH 6.5 | | 分子名称: | OXA10 beta-lactamase, SULFATE ION | | 著者 | Golemi, D, Maveyraud, L, Vakulenko, S, Samama, J.P, Mobashery, S. | | 登録日 | 2001-10-10 | | 公開日 | 2001-11-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Critical involvement of a carbamylated lysine in catalytic function of class D beta-lactamases.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
6O20
 
 | | Cryo-EM structure of TRPV5 with calmodulin bound | | 分子名称: | CALCIUM ION, Calmodulin, Transient receptor potential cation channel subfamily V member 5 | | 著者 | Dang, S, van Goor, M.K, Asarnow, D, Wang, Y, Julius, D, Cheng, Y, van der Wijst, J. | | 登録日 | 2019-02-22 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural insight into TRPV5 channel function and modulation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1KRX
 
 | | SOLUTION STRUCTURE OF BERYLLOFLUORIDE-ACTIVATED NTRC RECEIVER DOMAIN: MODEL STRUCTURES INCORPORATING ACTIVE SITE CONTACTS | | 分子名称: | BERYLLIUM TRIFLUORIDE ION, NITROGEN REGULATION PROTEIN NR(I) | | 著者 | Hastings, C.A, Lee, S.-Y, Cho, H.S, Yan, D, Kustu, S, Wemmer, D.E. | | 登録日 | 2002-01-10 | | 公開日 | 2003-08-19 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | High-Resolution Solution Structure of the Beryllofluoride-Activated NtrC Receiver Domain
Biochemistry, 42, 2003
|
|
1KRW
 
 | | SOLUTION STRUCTURE AND BACKBONE DYNAMICS OF BERYLLOFLUORIDE-ACTIVATED NTRC RECEIVER DOMAIN | | 分子名称: | NITROGEN REGULATION PROTEIN NR(I) | | 著者 | Hastings, C.A, Lee, S.-Y, Cho, H.S, Yan, D, Kustu, S, Wemmer, D.E. | | 登録日 | 2002-01-10 | | 公開日 | 2003-08-19 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | High-Resolution Solution Structure of the Beryllofluoride-Activated NtrC Receiver Domain
Biochemistry, 42, 2003
|
|
8HIU
 
 | |
8H8F
 
 | | Structure of Xenopus tropicalis acid-sensitive outwardly rectifying channel ASOR (resting state) | | 分子名称: | Proton-activated chloride channel | | 著者 | Chi, P, Wang, X, Li, J, Li, K, Zhang, Y, Geng, J, Wu, J, Deng, D. | | 登録日 | 2022-10-22 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-09-04 | | 実験手法 | ELECTRON MICROSCOPY (3.48 Å) | | 主引用文献 | Molecular insights into the inhibition of proton-activated chloride channel by transfer RNA.
Cell Res., 2024
|
|
8H8D
 
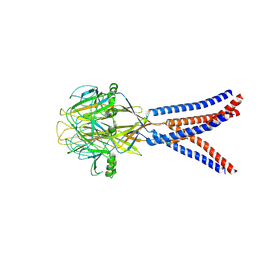 | | Structure of Xenopus tropicalis acid-sensitive outwardly rectifying channel ASOR trimer bound with tRNA (intermediate state) | | 分子名称: | Proton-activated chloride channel | | 著者 | Chi, P, Wang, X, Li, J, Li, K, Zhang, Y, Geng, J, Wu, J, Deng, D. | | 登録日 | 2022-10-22 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-09-04 | | 実験手法 | ELECTRON MICROSCOPY (4.26 Å) | | 主引用文献 | Molecular insights into the inhibition of proton-activated chloride channel by transfer RNA.
Cell Res., 2024
|
|
8H8E
 
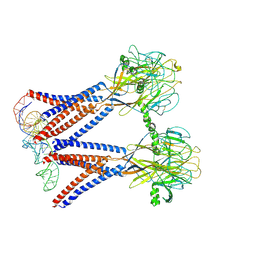 | | Structure of the dimeric Xenopus tropical acid-sensitive outwardly rectifying channel ASOR trimer bound with tRNA (closed state) | | 分子名称: | Proton-activated chloride channel, tRNA (75-MER)of Spodoptera frugiperda | | 著者 | Chi, P, Wang, X, Li, J, Li, K, Zhang, Y, Geng, J, Wu, J, Deng, D. | | 登録日 | 2022-10-22 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-09-04 | | 実験手法 | ELECTRON MICROSCOPY (3.81 Å) | | 主引用文献 | Molecular insights into the inhibition of proton-activated chloride channel by transfer RNA.
Cell Res., 2024
|
|
5LJM
 
 | | Structure of SPATA2 PUB domain | | 分子名称: | GLYCEROL, Spermatogenesis-associated protein 2 | | 著者 | Elliott, P.R, Komander, D. | | 登録日 | 2016-07-18 | | 公開日 | 2016-08-24 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.454 Å) | | 主引用文献 | SPATA2 Links CYLD to LUBAC, Activates CYLD, and Controls LUBAC Signaling.
Mol.Cell, 63, 2016
|
|
