5GIQ
 
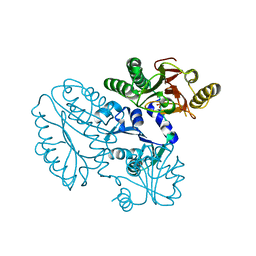 | | Xaa-Pro peptidase from Deinococcus radiodurans, Zinc bound | | 分子名称: | PHOSPHATE ION, Proline dipeptidase, ZINC ION | | 著者 | Are, V.N, Singh, R, Kumar, A, Ghosh, B, Jamdar, S.N, Makde, R.D. | | 登録日 | 2016-06-24 | | 公開日 | 2017-06-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structures and activities of widely conserved small prokaryotic aminopeptidases-P clarify classification of M24B peptidases.
Proteins, 2018
|
|
4EN2
 
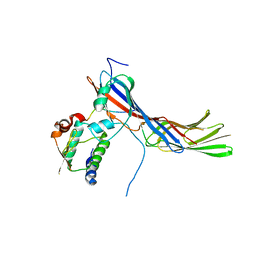 | |
4EMZ
 
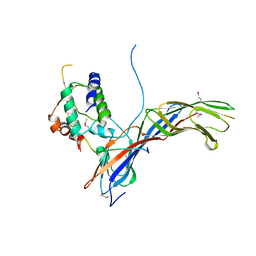 | |
3P14
 
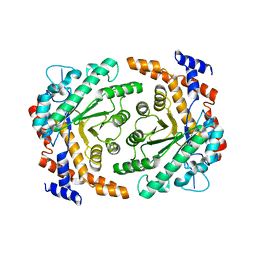 | |
2MV7
 
 | |
3QMW
 
 | |
3QMV
 
 | |
5GIV
 
 | | Crystal structure of M32 carboxypeptidase from Deinococcus radiodurans R1 | | 分子名称: | ACETATE ION, Carboxypeptidase 1, ZINC ION | | 著者 | Sharma, B, Singh, R, Yadav, P, Ghosh, B, Kumar, A, Jamdar, S.N, Makde, R.D. | | 登録日 | 2016-06-25 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Active site gate of M32 carboxypeptidases illuminated by crystal structure and molecular dynamics simulations
Biochim. Biophys. Acta, 1865, 2017
|
|
6A4R
 
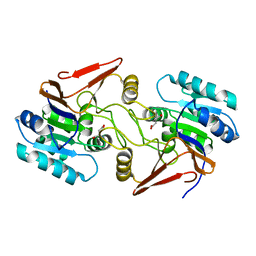 | | Crystal structure of aspartate bound peptidase E from Salmonella enterica | | 分子名称: | ASPARTIC ACID, Peptidase E | | 著者 | Yadav, P, Chandravanshi, K, Goyal, V.D, Singh, R, Kumar, A, Gokhale, S.M, Makde, R.D. | | 登録日 | 2018-06-20 | | 公開日 | 2018-10-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.828 Å) | | 主引用文献 | Structure of Asp-bound peptidase E from Salmonella enterica: Active site at dimer interface illuminates Asp recognition.
FEBS Lett., 592, 2018
|
|
6A4S
 
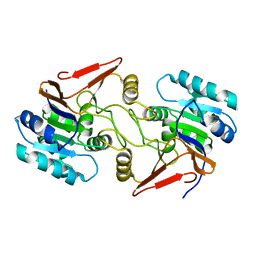 | | Crystal structure of peptidase E with ordered active site loop from Salmonella enterica | | 分子名称: | Peptidase E | | 著者 | Yadav, P, Chandravanshi, K, Goyal, V.D, Singh, R, Kumar, A, Gokhale, S.M, Makde, R.D. | | 登録日 | 2018-06-20 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of Asp-bound peptidase E from Salmonella enterica: Active site at dimer interface illuminates Asp recognition.
FEBS Lett., 592, 2018
|
|
2J7K
 
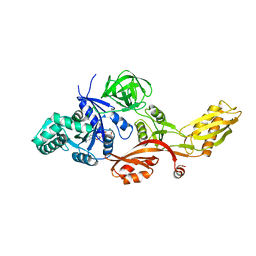 | |
1KQP
 
 | | NH3-DEPENDENT NAD+ SYNTHETASE FROM BACILLUS SUBTILIS AT 1 A RESOLUTION | | 分子名称: | 1,2-ETHANEDIOL, MAGNESIUM ION, NH(3)-dependent NAD(+) synthetase, ... | | 著者 | Symersky, J, Devedjiev, Y, Moore, K, Brouillette, C, DeLucas, L. | | 登録日 | 2002-01-07 | | 公開日 | 2002-06-28 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.03 Å) | | 主引用文献 | NH3-dependent NAD+ synthetase from Bacillus subtilis at 1 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
