5THE
 
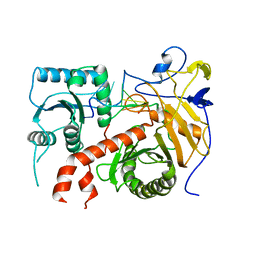 | |
7VHD
 
 | | Crystal structure of the STX2a complexed with R4A peptide | | 分子名称: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, ARG-ARG-ARG-ARG-ALA, Shiga toxin 2 B subunit, ... | | 著者 | Senda, M, Takahashi, M, Nishikawa, K, Senda, T. | | 登録日 | 2021-09-22 | | 公開日 | 2022-07-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A unique peptide-based pharmacophore identifies an inhibitory compound against the A-subunit of Shiga toxin.
Sci Rep, 12, 2022
|
|
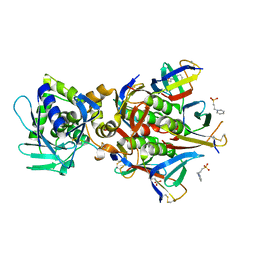
5VM9
 
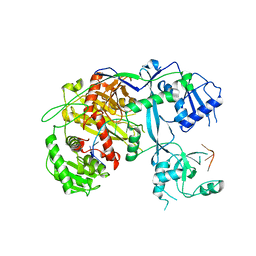 | | Human Argonaute3 bound to guide RNA | | 分子名称: | Protein argonaute-3, RNA (5'-R(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*UP*U)-3'), RNA (5'-R(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*UP*U)-3') | | 著者 | Park, M.S, Nakanishi, K. | | 登録日 | 2017-04-26 | | 公開日 | 2017-10-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.28 Å) | | 主引用文献 | Human Argonaute3 has slicer activity.
Nucleic Acids Res., 45, 2017
|
|
3AM2
 
 | | Clostridium perfringens enterotoxin | | 分子名称: | GLYCEROL, Heat-labile enterotoxin B chain, UNKNOWN ATOM OR ION | | 著者 | Kitadokoro, K, Nishimura, K, Kamitani, S, Kimura, J, Fukui, A, Abe, H, Horiguchi, Y. | | 登録日 | 2010-08-12 | | 公開日 | 2011-04-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Crystal Structure of Clostridium perfringens Enterotoxin Displays Features of {beta}-Pore-forming Toxins
J.Biol.Chem., 286, 2011
|
|
2ODO
 
 | |
7EBC
 
 | | Crystal structure of Isocitrate lyase-1 from Saccaromyces cervisiae | | 分子名称: | Isocitrate lyase, MAGNESIUM ION, TETRAETHYLENE GLYCOL | | 著者 | Hiragi, K, Nishio, K, Moriyama, S, Hamaguchi, T, Mizoguchi, A, Yonekura, K, Tani, K, Mizushima, T. | | 登録日 | 2021-03-09 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural insights into the targeting specificity of ubiquitin ligase for S. cerevisiae isocitrate lyase but not C. albicans isocitrate lyase.
J.Struct.Biol., 213, 2021
|
|
7EBF
 
 | | Cryo-EM structure of Isocitrate lyase-1 from Candida albicans | | 分子名称: | Isocitrate lyase | | 著者 | Hiragi, K, Nishio, K, Moriyama, S, Hamaguchi, T, Mizoguchi, A, Yonekura, K, Tani, K, Mizushima, T. | | 登録日 | 2021-03-09 | | 公開日 | 2021-06-23 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.63 Å) | | 主引用文献 | Structural insights into the targeting specificity of ubiquitin ligase for S. cerevisiae isocitrate lyase but not C. albicans isocitrate lyase.
J.Struct.Biol., 213, 2021
|
|
7EBE
 
 | | Crystal structure of Isocitrate lyase-1 from Candida albicans | | 分子名称: | FORMIC ACID, Isocitrate lyase, MAGNESIUM ION | | 著者 | Hiragi, K, Nishio, K, Moriyama, S, Hamaguchi, T, Mizoguchi, A, Yonekura, K, Tani, K, Mizushima, T. | | 登録日 | 2021-03-09 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Structural insights into the targeting specificity of ubiquitin ligase for S. cerevisiae isocitrate lyase but not C. albicans isocitrate lyase.
J.Struct.Biol., 213, 2021
|
|
5B37
 
 | | Crystal structure of L-tryptophan dehydrogenase from Nostoc punctiforme | | 分子名称: | Tryptophan dehydrogenase | | 著者 | Wakamatsu, T, Sakuraba, H, Kitamura, M, Hakumai, Y, Ohnishi, K, Ashiuchi, M, Ohshima, T. | | 登録日 | 2016-02-11 | | 公開日 | 2016-11-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Structural Insights into l-Tryptophan Dehydrogenase from a Photoautotrophic Cyanobacterium, Nostoc punctiforme.
Appl. Environ. Microbiol., 83, 2017
|
|
8GCL
 
 | | Cryo-EM structure of hAQP2 in DDM | | 分子名称: | Aquaporin-2 | | 著者 | Kamegawa, A, Suzuki, S, Nishikawa, K, Numoto, N, Suzuki, H, Fujiyoshi, Y. | | 登録日 | 2023-03-02 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.89 Å) | | 主引用文献 | Structural analysis of the water channel AQP2 by single-particle cryo-EM.
J.Struct.Biol., 215, 2023
|
|
3A57
 
 | | Crystal structure of Thermostable Direct Hemolysin | | 分子名称: | Thermostable direct hemolysin 2 | | 著者 | Hashimoto, H, Yanagihara, I, Nakahira, K, Hamada, D, Ikegami, T, Mayanagi, K, Kaieda, S, Fukui, T, Ohnishi, K, Kajiyama, S, Yamane, T, Ikeguchi, M, Honda, T, Shimizu, T, Sato, M. | | 登録日 | 2009-08-03 | | 公開日 | 2010-03-31 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure and functional characterization of Vibrio parahaemolyticus thermostable direct hemolysin
J.Biol.Chem., 285, 2010
|
|
4OM8
 
 | | Crystal structure of 5-formly-3-hydroxy-2-methylpyridine 4-carboxylic acid (FHMPC) 5-dehydrogenase, an NAD+ dependent dismutase. | | 分子名称: | 3-hydroxybutyryl-coA dehydrogenase, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | 著者 | Mugo, A.N, Kobayashi, J, Mikami, B, Yagi, T, Ohnishi, K. | | 登録日 | 2014-01-27 | | 公開日 | 2015-01-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal structure of 5-formyl-3-hydroxy-2-methylpyridine 4-carboxylic acid 5-dehydrogenase, an NAD(+)-dependent dismutase from Mesorhizobium loti
Biochem.Biophys.Res.Commun., 456, 2015
|
|
5ZI2
 
 | | MDH3 wild type, nad-form | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | 著者 | Moriyama, S, Nishio, K, Mizushima, T. | | 登録日 | 2018-03-14 | | 公開日 | 2018-10-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of glyoxysomal malate dehydrogenase (MDH3) from Saccharomyces cerevisiae.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5ZI3
 
 | | MDH3 wild type, apo-form | | 分子名称: | GLYCEROL, Malate dehydrogenase | | 著者 | Moriyama, S, Nishio, K, Mizushima, T. | | 登録日 | 2018-03-14 | | 公開日 | 2018-10-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of glyoxysomal malate dehydrogenase (MDH3) from Saccharomyces cerevisiae.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5ZI4
 
 | | MDH3 wild type, nad-oaa-form | | 分子名称: | Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXALOACETATE ION | | 著者 | Moriyama, S, Nishio, K, Mizushima, T. | | 登録日 | 2018-03-14 | | 公開日 | 2018-10-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of glyoxysomal malate dehydrogenase (MDH3) from Saccharomyces cerevisiae.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
7FI3
 
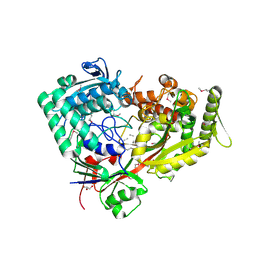 | | Archaeal oligopeptide permease A (OppA) from Thermococcus kodakaraensis in complex with an endogenous pentapeptide | | 分子名称: | ABC-type dipeptide/oligopeptide transport system, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | 著者 | Yokoyama, H, Kamei, N, Konishi, K, Hara, K, Hashimoto, H. | | 登録日 | 2021-07-30 | | 公開日 | 2022-04-13 | | 最終更新日 | 2022-06-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for peptide recognition by archaeal oligopeptide permease A.
Proteins, 90, 2022
|
|
5II4
 
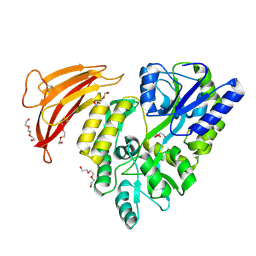 | | Crystal structure of red abalone VERL repeat 1 with linker at 2.0 A resolution | | 分子名称: | Maltose-binding periplasmic protein,Vitelline envelope sperm lysin receptor, TRIETHYLENE GLYCOL, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Sadat Al-Hosseini, H, Raj, I, Nishimura, K, Jovine, L. | | 登録日 | 2016-03-01 | | 公開日 | 2017-06-14 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
2Z1I
 
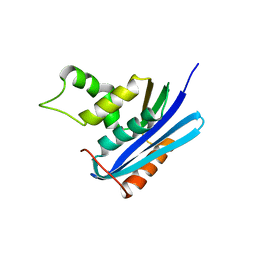 | | Crystal structure of E.coli RNase HI surface charged mutant(Q4R/T40E/Q72H/Q76K/Q80E/T92K/Q105K/Q113R/Q115K) | | 分子名称: | Ribonuclease HI | | 著者 | You, D.J, Fukuchi, S, Nishikawa, K, Koga, Y, Takano, K, Kanaya, S. | | 登録日 | 2007-05-08 | | 公開日 | 2007-11-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Protein Thermostabilization Requires a Fine-tuned Placement of Surface-charged Residues
J.Biochem.(Tokyo), 142, 2007
|
|
2Z1J
 
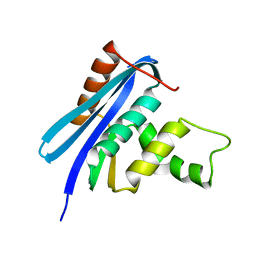 | | Crystal structure of E.coli RNase HI surface charged mutant(Q4R/T40E/Q72H/Q76K/Q80E/T92K/Q105K/Q113R/Q115K/N143K/T145K) | | 分子名称: | Ribonuclease HI | | 著者 | You, D.J, Fukuchi, S, Nishikawa, K, Koga, Y, Takano, K, Kanaya, S. | | 登録日 | 2007-05-08 | | 公開日 | 2007-11-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Protein Thermostabilization Requires a Fine-tuned Placement of Surface-charged Residues
J.Biochem.(Tokyo), 142, 2007
|
|
2Z1H
 
 | | Crystal structure of E.coli RNase HI surface charged mutant(Q4R/T92K/Q105K/Q113R/Q115K/N143K/T145K) | | 分子名称: | Ribonuclease HI | | 著者 | You, D.J, Fukuchi, S, Nishikawa, K, Koga, Y, Takano, K, Kanaya, S. | | 登録日 | 2007-05-08 | | 公開日 | 2007-11-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Protein Thermostabilization Requires a Fine-tuned Placement of Surface-charged Residues
J.Biochem.(Tokyo), 142, 2007
|
|
2Z1G
 
 | | Crystal structure of E.coli RNase HI surface charged mutant(Q4R/T40E/Q72H/Q76K/Q80E/T92K/Q105K) | | 分子名称: | Ribonuclease HI | | 著者 | You, D.J, Fukuchi, S, Nishikawa, K, Koga, Y, Takano, K, Kanaya, S. | | 登録日 | 2007-05-08 | | 公開日 | 2007-11-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Protein Thermostabilization Requires a Fine-tuned Placement of Surface-charged Residues
J.Biochem.(Tokyo), 142, 2007
|
|
2Z9W
 
 | | Crystal structure of pyridoxamine-pyruvate aminotransferase complexed with pyridoxal | | 分子名称: | 3-HYDROXY-5-(HYDROXYMETHYL)-2-METHYLISONICOTINALDEHYDE, Aspartate aminotransferase, GLYCEROL, ... | | 著者 | Yoshikane, Y, Yokochi, N, Yamasaki, M, Mizutani, K, Ohnishi, K, Mikami, B, Hayashi, H, Yagi, T. | | 登録日 | 2007-09-26 | | 公開日 | 2007-11-06 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti MAFF303099
J.Biol.Chem., 283, 2008
|
|
1EJ3
 
 | | CRYSTAL STRUCTURE OF AEQUORIN | | 分子名称: | AEQUORIN, C2-HYDROPEROXY-COELENTERAZINE | | 著者 | Head, J.F, Inouye, S, Teranishi, K, Shimomura, O. | | 登録日 | 2000-02-29 | | 公開日 | 2000-05-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The crystal structure of the photoprotein aequorin at 2.3 A resolution.
Nature, 405, 2000
|
|
3VIS
 
 | | Crystal structure of cutinase Est119 from Thermobifida alba AHK119 | | 分子名称: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Esterase | | 著者 | Kitadokoro, K, Thumarat, U, Nakamura, R, Nishimura, K, Karatani, H, Suzuki, H, Kawai, F. | | 登録日 | 2011-10-11 | | 公開日 | 2012-04-11 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Crystal structure of cutinase Est119 from Thermobida alba AHK119 that can degrade modpolyethylene terephthalate at 1.76 A resolution.
POLYM.DEGRAD.STAB., 97, 2012
|
|
3VVZ
 
 | | Crystal Strucuture of The Rhodamine 6G-Bound Form of RamR (Transcriptional Regurator of TetR Family) from Salmonella Typhimurium | | 分子名称: | Putative regulatory protein, RHODAMINE 6G, SULFATE ION | | 著者 | Sakurai, K, Nikaido, E, Nakashima, R, Yamasaki, S, Yamaguchi, A, Nishino, K. | | 登録日 | 2012-07-30 | | 公開日 | 2013-07-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | The crystal structure of multidrug-resistance regulator RamR with multiple drugs
Nat Commun, 4, 2013
|
|
