3MX2
 
 | | Lassa fever virus Nucleoprotein complexed with dTTP | | 分子名称: | Nucleoprotein, THYMIDINE-5'-TRIPHOSPHATE, ZINC ION | | 著者 | Qi, X, Lan, S, Wang, W, Schelde, L.M, Dong, H, Wallat, G, Liang, Y, Ly, H, Dong, C, Scottish Structural Proteomics Facility (SSPF) | | 登録日 | 2010-05-06 | | 公開日 | 2010-12-01 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.983 Å) | | 主引用文献 | Cap binding and immune evasion revealed by Lassa nucleoprotein structure.
Nature, 468, 2010
|
|
4URK
 
 | | PI3Kg in complex with AZD6482 | | 分子名称: | 2-[[(1R)-1-(7-methyl-2-morpholin-4-yl-4-oxidanylidene-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid, PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | 著者 | Giordanetto, F, Barlaam, B, Berglund, S, Edman, K, Karlsson, O, Lindberg, J, Nylander, S, Inghardt, T. | | 登録日 | 2014-06-30 | | 公開日 | 2014-10-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Discovery of 9-(1-Phenoxyethyl)-2-Morpholino-4-Oxo-Pyrido[1, 2-A]Pyrimidine-7-Carboxamides as Oral Pi3Kbeta Inhibitors, Useful as Antiplatelet Agents.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4V96
 
 | | The structure of a 1.8 MDa viral genome injection device suggests alternative infection mechanisms | | 分子名称: | BPP, ORF46, ORF48 | | 著者 | Veesler, D, Spinelli, S, Mahony, J, Lichiere, J, Blangy, S, Bricogne, G, Legrand, P, Ortiz-Lombardia, M, Campanacci, V, van Sinderen, D, Cambillau, C. | | 登録日 | 2012-02-01 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4IOS
 
 | | Structure of phage TP901-1 RBP (ORF49) in complex with nanobody 11. | | 分子名称: | BPP, GLYCEROL, Llama nanobody 11 | | 著者 | Desmyter, A, Farenc, C, Mahony, J, Spinelli, S, Bebeacua, C, Blangy, S, Veesler, D, van Sinderen, D, Cambillau, C. | | 登録日 | 2013-01-08 | | 公開日 | 2013-03-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Viral infection modulation and neutralization by camelid nanobodies
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
9EZ8
 
 | | Cryo-EM structure of the icosahedral lumazine synthase from Vicia faba. | | 分子名称: | 6,7-dimethyl-8-ribityllumazine synthase | | 著者 | Chee, M, Trapani, S, Hoh, F, Lai Kee Him, J, Yvon, M, Blanc, S, Bron, P. | | 登録日 | 2024-04-11 | | 公開日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Cryo-EM structure of the icosahedral lumazine synthase from Vicia faba.
To Be Published
|
|
4IPT
 
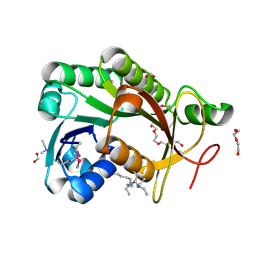 | | The crystal structure of a short-chain dehydrogenases/reductase (ethylated) from Veillonella parvula DSM 2008 | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | 著者 | Tan, K, Hatzos-Skintges, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2013-01-10 | | 公開日 | 2013-02-06 | | 最終更新日 | 2019-11-27 | | 実験手法 | X-RAY DIFFRACTION (1.546 Å) | | 主引用文献 | The crystal structure of a short-chain dehydrogenases/reductase (ethylated) from Veillonella parvula DSM 2008
To be Published
|
|
4W9R
 
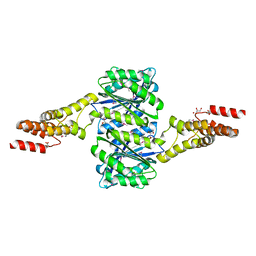 | | Crystal structure of uncharacterised protein Coch_1243 from Capnocytophaga ochracea DSM 7271 | | 分子名称: | ACETATE ION, GLYCEROL, Uncharacterized protein | | 著者 | Chang, C, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-08-27 | | 公開日 | 2014-09-10 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.703 Å) | | 主引用文献 | Crystal structure of uncharacterised protein Coch_1243 from Capnocytophaga ochracea DSM 7271
To Be Published
|
|
4UQX
 
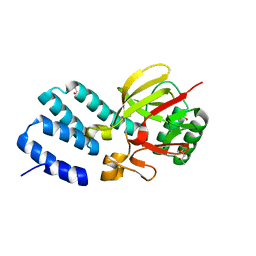 | | Coevolution of the ATPase ClpV, the TssB-TssC Sheath and the Accessory HsiE Protein Distinguishes Two Type VI Secretion Classes | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, HSIE1 | | 著者 | Forster, A, Planamente, S, Manoli, E, Lossi, N.S, Freemont, P.S, Filloux, A. | | 登録日 | 2014-06-25 | | 公開日 | 2014-10-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Coevolution of the ATPase Clpv, the Sheath Proteins Tssb and Tssc and the Accessory Protein Tagj/Hsie1 Distinguishes Type Vi Secretion Classes.
J.Biol.Chem., 289, 2014
|
|
4UQW
 
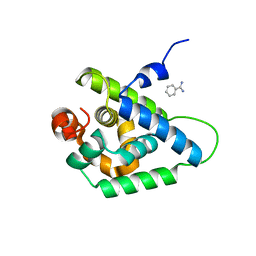 | | Coevolution of the ATPase ClpV, the TssB-TssC Sheath and the Accessory HsiE Protein Distinguishes Two Type VI Secretion Classes | | 分子名称: | BENZAMIDINE, PROTEIN CLPV1 | | 著者 | Forster, A, Planamente, S, Manoli, E, Lossi, N.S, Freemont, P.S, Filloux, A. | | 登録日 | 2014-06-25 | | 公開日 | 2014-10-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Coevolution of the ATPase Clpv, the Sheath Proteins Tssb and Tssc and the Accessory Protein Tagj/Hsie1 Distinguishes Type Vi Secretion Classes.
J.Biol.Chem., 289, 2014
|
|
5UQP
 
 | | The crystal structure of cupin protein from Rhodococcus jostii RHA1 | | 分子名称: | CHLORIDE ION, Cupin, SULFATE ION, ... | | 著者 | Tan, K, Li, H, Clancy, S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2017-02-08 | | 公開日 | 2017-02-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The crystal structure of cupin protein from Rhodococcus jostii RHA1
To Be Published
|
|
3EA0
 
 | | Crystal Structure of ParA Family ATPase from Chlorobium tepidum TLS | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, ATPase, ParA family, ... | | 著者 | Kim, Y, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-08-24 | | 公開日 | 2008-09-23 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of ParA Family ATPase from Chlorobium tepidum TLS
To be Published
|
|
3EAG
 
 | |
3ERM
 
 | |
3EY5
 
 | | Putative acetyltransferase from GNAT family from Bacteroides thetaiotaomicron. | | 分子名称: | Acetyltransferase-like, GNAT family, SULFATE ION | | 著者 | Osipiuk, J, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-10-17 | | 公開日 | 2008-11-18 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | X-ray crystal structure of putative acetyltransferase from GNAT family from Bacteroides thetaiotaomicron.
To be Published
|
|
3F0A
 
 | | Structure of a putative n-acetyltransferase (ta0374) in complex with acetyl-coa from thermoplasma acidophilum | | 分子名称: | ACETYL COENZYME *A, CHLORIDE ION, N-ACETYLTRANSFERASE, ... | | 著者 | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-10-24 | | 公開日 | 2008-11-11 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of the novel PaiA N-acetyltransferase from Thermoplasma acidophilum involved in the negative control of sporulation and degradative enzyme production.
Proteins, 79, 2011
|
|
4RGA
 
 | | Phage 1358 receptor binding protein in complex with the trisaccharide GlcNAc-Galf-GlcOMe | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-galactofuranose-(1-6)-methyl alpha-D-glucopyranoside, Phage 1358 receptor binding protein (ORF20) | | 著者 | Spinelli, S, Mccabe, O, Farenc, C, Tremblay, D, Blangy, S, Oscarson, S, Moineau, S, Cambillau, C. | | 登録日 | 2014-09-29 | | 公開日 | 2015-05-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The targeted recognition of Lactococcus lactis phages to their polysaccharide receptors.
Mol.Microbiol., 96, 2015
|
|
4RA7
 
 | | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with nafcillin | | 分子名称: | (2R,4S)-2-[(1R)-2-hydroxy-1-{[(2-hydroxynaphthalen-1-yl)carbonyl]amino}ethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Peptidoglycan glycosyltransferase | | 著者 | Filippova, E.V, Minasov, G, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-09-09 | | 公開日 | 2014-09-24 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with nafcillin
To be Published
|
|
4R1H
 
 | |
4RD7
 
 | | The crystal structure of a Cupin 2 conserved barrel domain protein from Salinispora arenicola CNS-205 | | 分子名称: | Cupin 2 conserved barrel domain protein, GLYCEROL, SULFATE ION | | 著者 | Tan, K, Gu, M, Clancy, S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2014-09-18 | | 公開日 | 2014-10-01 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.571 Å) | | 主引用文献 | The crystal structure of a Cupin 2 conserved barrel domain protein from Salinispora arenicola CNS-205
To be Published
|
|
4RGP
 
 | | Crystal Structure of Uncharacterized CRISPR/Cas System-associated Protein Csm6 from Streptococcus mutans | | 分子名称: | CALCIUM ION, Csm6_III-A, GLYCEROL, ... | | 著者 | Kim, Y, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-09-30 | | 公開日 | 2014-12-24 | | 実験手法 | X-RAY DIFFRACTION (2.299 Å) | | 主引用文献 | Crystal Structure of Uncharacterized CRISPR/Cas System-associated Protein Csm6 from Streptococcus mutans
To be Published
|
|
4R23
 
 | | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with dicloxacillin | | 分子名称: | (3R,4R,5R)-3-(2,6-dichlorophenyl)-N-{(1R)-1-[(2R,4S)-4-(dihydroxymethyl)-5,5-dimethyl-1,3-thiazolidin-2-yl]-2-oxoethyl} -5-methyl-1,2-oxazolidine-4-carboxamide, 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, ... | | 著者 | Filippova, E.V, Minasov, G, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-08-08 | | 公開日 | 2014-09-17 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with dicloxacillin
To be Published
|
|
4RHH
 
 | | Crystal structure of the catalytic mutant Xyn52B2-E335G, a GH52 Beta-D-xylosidase from Geobacillus stearothermophilus T6 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-xylosidase, CALCIUM ION | | 著者 | Dann, R, Lansky, S, Lavid, N, Zehavi, A, Belakhov, V, Baasov, T, Manjasetty, B, Belrhali, H, Shoham, Y, Shoham, G. | | 登録日 | 2014-10-02 | | 公開日 | 2015-10-14 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | To be published
To be Published
|
|
4RJ0
 
 | |
4RIZ
 
 | |
4RIT
 
 | | The yellow crystal structure of pyridoxal-dependent decarboxylase from sphaerobacter thermophilus dsm 20745 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | 著者 | Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-10-07 | | 公開日 | 2014-10-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The yellow crystal structure of pyridoxal-dependent decarboxylase from sphaerobacter thermophilus dsm 20745
To be Published
|
|
