6LIY
 
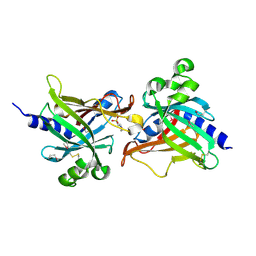 | | SeMet CRL Protein of Arabidopsis | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Chromophore lyase CRL, chloroplastic | | 著者 | Wang, F.F, Guan, K.L, Sun, P.K, Xing, W.M. | | 登録日 | 2019-12-13 | | 公開日 | 2020-09-16 | | 最終更新日 | 2020-12-02 | | 実験手法 | X-RAY DIFFRACTION (1.761 Å) | | 主引用文献 | The Arabidopsis CRUMPLED LEAF protein, a homolog of the cyanobacterial bilin lyase, retains the bilin-binding pocket for a yet unknown function.
Plant J., 104, 2020
|
|
5BV6
 
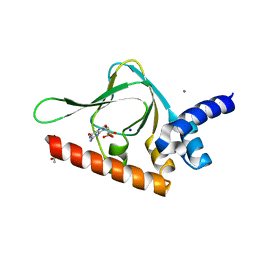 | | PKG II's Carboxyl Terminal Cyclic Nucleotide Binding Domain (CNB-B) in a complex with cGMP | | 分子名称: | ACETATE ION, CALCIUM ION, GUANOSINE-3',5'-MONOPHOSPHATE, ... | | 著者 | Campbell, J.C, Reger, A.S, Huang, G.Y, Sankaran, B, Kim, J.J, Kim, C.W. | | 登録日 | 2015-06-04 | | 公開日 | 2016-01-20 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Structural Basis of Cyclic Nucleotide Selectivity in cGMP-dependent Protein Kinase II.
J.Biol.Chem., 291, 2016
|
|
5X90
 
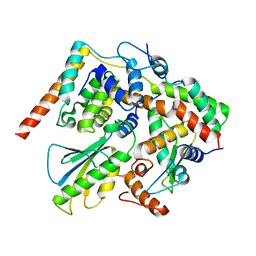 | | Structure of DotL(656-783)-IcmS-IcmW-LvgA derived from Legionella pneumophila | | 分子名称: | Hypothetical virulence protein, IcmO (DotL), IcmS, ... | | 著者 | Kim, H, Kwak, M.J, Kim, J.D, Kim, Y.G, Oh, B.H. | | 登録日 | 2017-03-04 | | 公開日 | 2017-06-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Architecture of the type IV coupling protein complex of Legionella pneumophila
Nat Microbiol, 2, 2017
|
|
7QA1
 
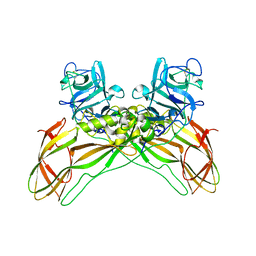 | | The structure of natural crystals of the Lysinibacillus sphaericus Tpp49Aa1 pesticidal protein elucidated using serial femtosecond crystallography at an X-ray free electron laser | | 分子名称: | Toxin-10 pesticidal protein (Tpp) 49Aa1 | | 著者 | Williamson, L.J, Rizkallah, P.J, Berry, C, Oberthur, D, Galchenkova, M, Yefanov, O, Bean, R, Best, H.L. | | 登録日 | 2021-11-15 | | 公開日 | 2023-05-17 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of the Lysinibacillus sphaericus Tpp49Aa1 pesticidal protein elucidated from natural crystals using MHz-SFX.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5VLX
 
 | | Dehaloperoxidase B mutant F21W | | 分子名称: | Dehaloperoxidase B, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Carey, L.M, Ghiladi, R.A. | | 登録日 | 2017-04-26 | | 公開日 | 2017-12-27 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Selective tuning of activity in a multifunctional enzyme as revealed in the F21W mutant of dehaloperoxidase B from Amphitrite ornata.
J. Biol. Inorg. Chem., 23, 2018
|
|
5CUF
 
 | | X-ray crystal structure of SeMet human Sestrin2 | | 分子名称: | Sestrin-2 | | 著者 | Kim, H, An, S, Ro, S.-H, Lee, J.H, Cho, U.-S. | | 登録日 | 2015-07-24 | | 公開日 | 2016-01-13 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Janus-faced Sestrin2 controls ROS and mTOR signalling through two separate functional domains.
Nat Commun, 6, 2015
|
|
5C6C
 
 | | PKG II's Amino Terminal Cyclic Nucleotide Binding Domain (CNB-A) in a complex with cAMP | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CADMIUM ION, ... | | 著者 | Campbell, J.C, Reger, A.S, Huang, G.Y, Sankaran, B, Kim, J.J, Kim, C.W. | | 登録日 | 2015-06-22 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural Basis of Cyclic Nucleotide Selectivity in cGMP-dependent Protein Kinase II.
J.Biol.Chem., 291, 2016
|
|
5C8W
 
 | | PKG II's Amino Terminal Cyclic Nucleotide Binding Domain (CNB-A) in a complex with cGMP | | 分子名称: | CYCLIC GUANOSINE MONOPHOSPHATE, MALONIC ACID, SODIUM ION, ... | | 著者 | Campbell, J.C, Reger, A.S, Huang, G.Y, Sankaran, B, Kim, J.J, Kim, C.W. | | 登録日 | 2015-06-26 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural Basis of Cyclic Nucleotide Selectivity in cGMP-dependent Protein Kinase II.
J.Biol.Chem., 291, 2016
|
|
7MBJ
 
 | |
7D01
 
 | |
7D02
 
 | |
7D04
 
 | |
7D05
 
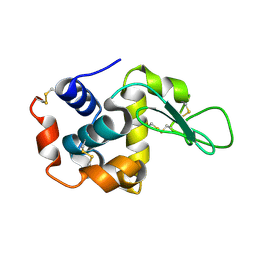 | |
7LV3
 
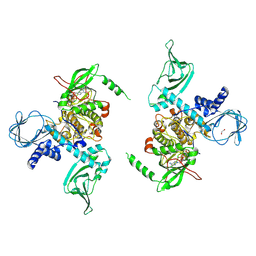 | | Crystal structure of human protein kinase G (PKG) R-C complex in inhibited state | | 分子名称: | 1,2-ETHANEDIOL, Isoform Beta of cGMP-dependent protein kinase 1, MANGANESE (II) ION, ... | | 著者 | Sharma, R, Lying, Q, Casteel, D, Kim, C. | | 登録日 | 2021-02-23 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | An auto-inhibited state of protein kinase G and implications for selective activation.
Elife, 11, 2022
|
|
4RSI
 
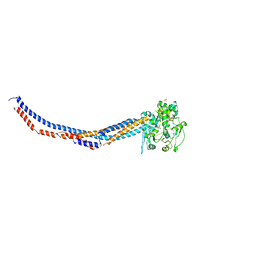 | |
4RSJ
 
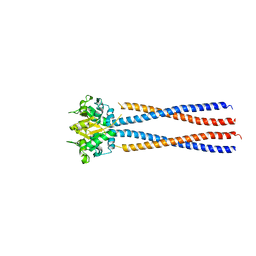 | |
1GUB
 
 | | Hinge-bending motion of D-allose binding protein from Escherichia coli: three open conformations | | 分子名称: | D-ALLOSE-BINDING PERIPLASMIC PROTEIN, NICKEL (II) ION | | 著者 | Magnusson, U, Chaudhuri, B.N, Ko, J, Park, C, Jones, T.A, Mowbray, S.L. | | 登録日 | 2002-01-24 | | 公開日 | 2003-03-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structure of D-Allose Binding Protein from Escherichia Coli Bound to D-Allose at 1.8 A Resolution
J.Mol.Biol., 286, 1999
|
|
1GUD
 
 | | Hinge-bending motion of D-allose binding protein from Escherichia coli: three open conformations | | 分子名称: | D-ALLOSE-BINDING PERIPLASMIC PROTEIN, ZINC ION | | 著者 | Magnusson, U, Chaudhuri, B.N, Ko, J, Park, C, Jones, T.A, Mowbray, S.L. | | 登録日 | 2002-01-24 | | 公開日 | 2003-03-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Structure of D-Allose Binding Protein from Escherichia Coli Bound to D-Allose at 1.8 A Resolution
J.Mol.Biol., 286, 1999
|
|
7E65
 
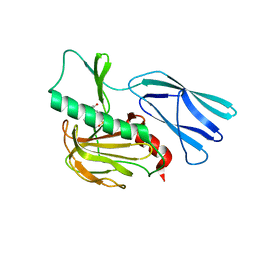 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 3 | | 分子名称: | (2S)-2-acetamido-N-[(2R)-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]-3-(4-sulfamoylphenyl)propanamide, Peptidase M23, ZINC ION | | 著者 | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | 登録日 | 2021-02-21 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E60
 
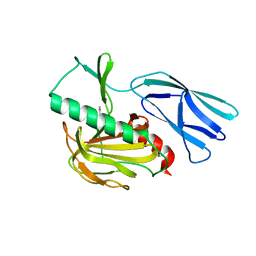 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 1 | | 分子名称: | (2~{R},6~{S})-2,6-diacetamido-7-[[(2~{R})-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]amino]-7-oxidanylidene-heptanoic acid, Peptidase M23, ZINC ION | | 著者 | Min, K, Yoon, H.J, Choi, Y, Lee, H.H. | | 登録日 | 2021-02-21 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E69
 
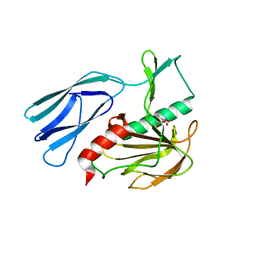 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 3-3 | | 分子名称: | N-oxidanyl-4-[(4-sulfamoylphenyl)methyl]benzamide, Peptidase M23, ZINC ION | | 著者 | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | 登録日 | 2021-02-21 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E61
 
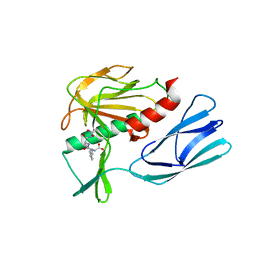 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 2 | | 分子名称: | 2-[[(3S)-3-acetamido-4-[[(2R)-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]amino]-4-oxidanylidene-butyl]-(phenylmethyl)amino]ethanoic acid, Peptidase M23, ZINC ION | | 著者 | Min, K.J, Yoon, H.J, Choi, Y, Lee, H.H. | | 登録日 | 2021-02-21 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E64
 
 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 2-2 | | 分子名称: | 2-[[(3S)-3-acetamido-4-[[(2R)-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]amino]-4-oxidanylidene-butyl]amino]ethanoic acid, Peptidase M23, ZINC ION | | 著者 | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | 登録日 | 2021-02-21 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E67
 
 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 3-2 | | 分子名称: | N-oxidanyl-2-[4-(4-sulfamoylphenyl)phenyl]ethanamide, Peptidase M23, ZINC ION | | 著者 | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | 登録日 | 2021-02-21 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E66
 
 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 3-1 | | 分子名称: | N-[2-(oxidanylamino)-2-oxidanylidene-ethyl]-2-(4-sulfamoylphenyl)ethanamide, Peptidase M23, ZINC ION | | 著者 | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | 登録日 | 2021-02-21 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
