4RQ9
 
 | | Crystal structure of the chromophore-binding domain of Stigmatella aurantiaca bacteriophytochrome (Thr289His mutant) in the Pr state | | 分子名称: | 1,2-ETHANEDIOL, BILIVERDINE IX ALPHA, GLYCEROL, ... | | 著者 | Woitowich, N.C, Halavaty, A.S, Gallagher, K.D, Nugent, A.C, Patel, H, Duong, P, Kovaleva, S.E, St.Peter, S, Ozarowski, W.B, Hernandez, C.N, Stojkovic, E.A. | | 登録日 | 2014-10-31 | | 公開日 | 2016-05-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of the chromophore-binding domain of Stigmatella aurantiaca bacteriophytochrome (Thr289His mutant) in the Pr state
To be Published
|
|
3TFC
 
 | | 1.95 Angstrom crystal structure of a bifunctional 3-deoxy-7-phosphoheptulonate synthase/chorismate mutase (aroA) from Listeria monocytogenes EGD-e in complex with phosphoenolpyruvate | | 分子名称: | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase, CHLORIDE ION, MANGANESE (II) ION, ... | | 著者 | Light, S.H, Minasov, G, Halavaty, A.S, Shuvalova, L, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-08-15 | | 公開日 | 2011-08-31 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural analysis of a 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase with an N-terminal chorismate mutase-like regulatory domain.
Protein Sci., 21, 2012
|
|
3ROI
 
 | | 2.20 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii | | 分子名称: | 3-phosphoshikimate 1-carboxyvinyltransferase, CHLORIDE ION, SULFATE ION | | 著者 | Light, S.H, Minasov, G, Krishna, S.N, Halavaty, A.S, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-04-25 | | 公開日 | 2011-05-18 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | 2.20 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii
To be Published
|
|
4NOG
 
 | | Crystal structure of a putative ornithine aminotransferase from Toxoplasma gondii ME49 in complex with pyrodoxal-5'-phosphate | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | 著者 | Filippova, E.V, Halavaty, A, Ruan, J, Shuvalova, L, Flores, K, Dubrovska, I, Ngo, H, Shanmugam, D, Roos, D, Anderson, W.F, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-11-19 | | 公開日 | 2013-12-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
4ECM
 
 | | 2.3 Angstrom Crystal Structure of a Glucose-1-phosphate Thymidylyltransferase from Bacillus anthracis in Complex with Thymidine-5-diphospho-alpha-D-glucose and Pyrophosphate | | 分子名称: | 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, Glucose-1-phosphate thymidylyltransferase, PYROPHOSPHATE 2- | | 著者 | Minasov, G, Kuhn, M, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2012-03-26 | | 公開日 | 2012-04-04 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of the Bacillus anthracis dTDP-L-rhamnose-biosynthetic enzyme glucose-1-phosphate thymidylyltransferase (RfbA).
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
6BAF
 
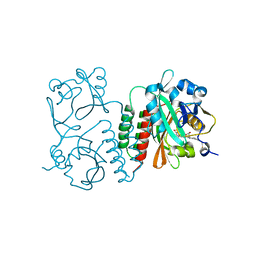 | | Structure of the chromophore binding domain of Stigmatella aurantiaca phytochrome P1, wild-type | | 分子名称: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | 著者 | Schmidt, M, Stojkovic, E. | | 登録日 | 2017-10-12 | | 公開日 | 2018-09-19 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural basis for light control of cell development revealed by crystal structures of a myxobacterial phytochrome.
IUCrJ, 5, 2018
|
|
6BAP
 
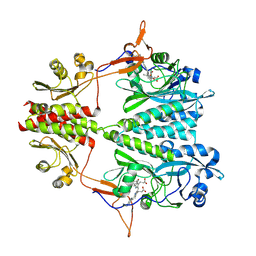 | | Stigmatella aurantiaca bacterial phytochrome PAS-GAF-PHY, T289H mutant | | 分子名称: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | 著者 | Schmidt, M, Stojkovic, E. | | 登録日 | 2017-10-14 | | 公開日 | 2018-09-19 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structural basis for light control of cell development revealed by crystal structures of a myxobacterial phytochrome.
IUCrJ, 5, 2018
|
|
6BAY
 
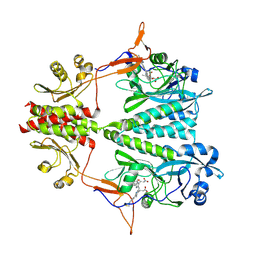 | |
6BAO
 
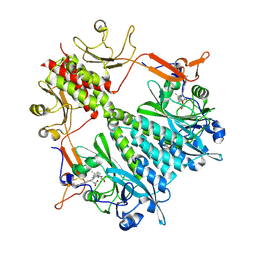 | | Stigmatella aurantiaca phytochrome photosensory core module, wild type | | 分子名称: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | 著者 | Schmidt, M, Stojkovic, E. | | 登録日 | 2017-10-14 | | 公開日 | 2018-09-19 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Structural basis for light control of cell development revealed by crystal structures of a myxobacterial phytochrome.
IUCrJ, 5, 2018
|
|
6BAK
 
 | | The structure of the Stigmatella aurantiaca phytochrome chromophore binding domain T289H mutant | | 分子名称: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | 著者 | Schmidt, M, Stojkovic, E. | | 登録日 | 2017-10-13 | | 公開日 | 2018-09-19 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structural basis for light control of cell development revealed by crystal structures of a myxobacterial phytochrome.
IUCrJ, 5, 2018
|
|
4NU7
 
 | | 2.05 Angstrom Crystal Structure of Ribulose-phosphate 3-epimerase from Toxoplasma gondii. | | 分子名称: | CHLORIDE ION, Ribulose-phosphate 3-epimerase, SULFATE ION, ... | | 著者 | Minasov, G, Ruan, J, Ngo, H, Shuvalova, L, Dubrovska, I, Flores, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-12-03 | | 公開日 | 2013-12-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
4ODI
 
 | | 2.6 Angstrom Crystal Structure of Putative Phosphoglycerate Mutase 1 from Toxoplasma gondii | | 分子名称: | Phosphoglycerate mutase PGMII, SODIUM ION | | 著者 | Minasov, G, Ruan, J, Ngo, H, Shuvalova, L, Dubrovska, I, Flores, K, Shanmugam, D, Roos, D, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-01-10 | | 公開日 | 2014-01-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
4O0N
 
 | | 2.4 Angstrom Resolution Crystal Structure of Putative Nucleoside Diphosphate Kinase from Toxoplasma gondii. | | 分子名称: | Nucleoside diphosphate kinase, SULFATE ION | | 著者 | Minasov, G, Ruan, J, Ngo, H, Shuvalova, L, Dubrovska, I, Flores, K, Shanmugam, D, Roos, D, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-12-13 | | 公開日 | 2013-12-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
5F7P
 
 | |
5F7V
 
 | |
5F7U
 
 | |
5F7R
 
 | |
5F7Q
 
 | |
5BXI
 
 | | 1.7 Angstrom Resolution Crystal Structure of Putative Nucleoside Diphosphate Kinase from Toxoplasma gondii with Tyrosine of Tag Bound to Active Site | | 分子名称: | BICARBONATE ION, DI(HYDROXYETHYL)ETHER, Nucleoside diphosphate kinase | | 著者 | Minasov, G, Ruan, J, Ngo, H, Shuvalova, L, Dubrovska, I, Flores, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-06-09 | | 公開日 | 2015-06-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
5F7S
 
 | |
5I0E
 
 | | Cycloalternan-degrading enzyme from Trueperella pyogenes in complex with isomaltose | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Glycoside hydrolase family 31, ... | | 著者 | Light, S.H, Minasov, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-02-03 | | 公開日 | 2016-12-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Transferase Versus Hydrolase: The Role of Conformational Flexibility in Reaction Specificity.
Structure, 25, 2017
|
|
5I0F
 
 | | Cycloalternan-degrading enzyme from Trueperella pyogenes in complex with covalent intermediate | | 分子名称: | CALCIUM ION, Glycoside hydrolase family 31, TRIETHYLENE GLYCOL, ... | | 著者 | Light, S.H, Minasov, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-02-03 | | 公開日 | 2016-12-14 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Transferase Versus Hydrolase: The Role of Conformational Flexibility in Reaction Specificity.
Structure, 25, 2017
|
|
5I0D
 
 | | Cycloalternan-forming enzyme from Listeria monocytogenes in complex with cycloalternan | | 分子名称: | CALCIUM ION, CHLORIDE ION, Cyclic alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose, ... | | 著者 | Light, S.H, Minasov, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-02-03 | | 公開日 | 2016-12-14 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Transferase Versus Hydrolase: The Role of Conformational Flexibility in Reaction Specificity.
Structure, 25, 2017
|
|
5I0G
 
 | | Cycloalternan-degrading enzyme from Trueperella pyogenes in complex with cycloalternan | | 分子名称: | Cyclic alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose, Glycoside hydrolase family 31, SUCCINIC ACID | | 著者 | Light, S.H, Minasov, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-02-03 | | 公開日 | 2016-12-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Transferase Versus Hydrolase: The Role of Conformational Flexibility in Reaction Specificity.
Structure, 25, 2017
|
|
4E0C
 
 | | 1.8 Angstrom Resolution Crystal Structure of Transaldolase from Francisella tularensis (phosphate-free) | | 分子名称: | ACETATE ION, MAGNESIUM ION, Transaldolase | | 著者 | Light, S.H, Minasov, G, Halavaty, A.S, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2012-03-02 | | 公開日 | 2012-03-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Adherence to Burgi-Dunitz stereochemical principles requires significant structural rearrangements in Schiff-base formation: insights from transaldolase complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
