8F9M
 
 | |
8F9G
 
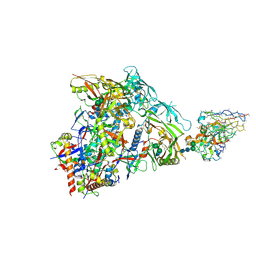 | | HIV Env germline targeting BG505_MD64_N332-GT5 SOSIP in complex with V3-glycan polyclonal Fab isolated from immunized BG18HCgl knock-in mice | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505_MD64_N332-GT5 gp120, ... | | 著者 | Ozorowski, G, Torres, J.L, Ward, A.B. | | 登録日 | 2022-11-23 | | 公開日 | 2024-05-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | mRNA-LNP HIV-1 trimer boosters elicit precursors to broad neutralizing antibodies.
Science, 384, 2024
|
|
3QAM
 
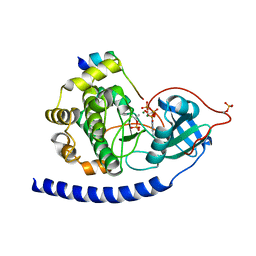 | | Crystal Structure of Glu208Ala mutant of catalytic subunit of cAMP-dependent protein kinase | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein kinase inhibitor, ... | | 著者 | Yang, J, Wu, J, Steichen, J, Taylor, S.S. | | 登録日 | 2011-01-11 | | 公開日 | 2011-12-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | A conserved Glu-Arg salt bridge connects coevolved motifs that define the eukaryotic protein kinase fold.
J.Mol.Biol., 415, 2012
|
|
3QAL
 
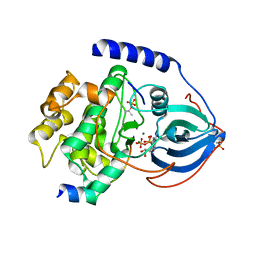 | | Crystal Structure of Arg280Ala mutant of Catalytic subunit of cAMP-dependent Protein Kinase | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein kinase inhibitor, ... | | 著者 | Yang, J, Wu, J, Steichen, J, Taylor, S.S. | | 登録日 | 2011-01-11 | | 公開日 | 2011-12-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | A conserved Glu-Arg salt bridge connects coevolved motifs that define the eukaryotic protein kinase fold.
J.Mol.Biol., 415, 2012
|
|
3NAD
 
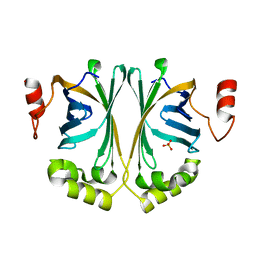 | | Crystal Structure of Phenolic Acid Decarboxylase from Bacillus pumilus UI-670 | | 分子名称: | Ferulate decarboxylase, SULFATE ION | | 著者 | Matte, A, Grosse, S, Bergeron, H, Abokitse, K, Lau, P.C.K. | | 登録日 | 2010-06-01 | | 公開日 | 2010-11-10 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Structural analysis of Bacillus pumilus phenolic acid decarboxylase, a lipocalin-fold enzyme.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
