3G0W
 
 | |
4JWH
 
 | | Crystal structure of spTrm10(Full length)-SAH complex | | 分子名称: | S-ADENOSYL-L-HOMOCYSTEINE, tRNA (guanine(9)-N1)-methyltransferase | | 著者 | Yan, W, Shao, Z. | | 登録日 | 2013-03-27 | | 公開日 | 2013-10-16 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Crystal structure of tRNA m1G9 methyltransferase Trm10: insight into the catalytic mechanism and recognition of tRNA substrate.
Nucleic Acids Res., 42, 2014
|
|
4JWJ
 
 | | Crystal structure of scTrm10(84)-SAH complex | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | 著者 | Yan, W, Shao, Z. | | 登録日 | 2013-03-27 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Crystal structure of tRNA m1G9 methyltransferase Trm10: insight into the catalytic mechanism and recognition of tRNA substrate.
Nucleic Acids Res., 42, 2014
|
|
4JWG
 
 | | Crystal structure of spTrm10(74) | | 分子名称: | ACETIC ACID, tRNA (guanine(9)-N1)-methyltransferase | | 著者 | Yan, W, Shao, Z. | | 登録日 | 2013-03-27 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of tRNA m1G9 methyltransferase Trm10: insight into the catalytic mechanism and recognition of tRNA substrate.
Nucleic Acids Res., 42, 2014
|
|
4JWF
 
 | | Crystal structure of spTrm10(74)-SAH complex | | 分子名称: | ACETIC ACID, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (guanine(9)-N1)-methyltransferase | | 著者 | Yan, W, Shao, Z. | | 登録日 | 2013-03-27 | | 公開日 | 2013-10-16 | | 最終更新日 | 2014-02-12 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of tRNA m1G9 methyltransferase Trm10: insight into the catalytic mechanism and recognition of tRNA substrate.
Nucleic Acids Res., 42, 2014
|
|
8ABV
 
 | | Crystal structure of SpLdpA in complex with erythro-DGPD | | 分子名称: | (1R,2S)-1,2-bis(3-methoxy-4-oxidanyl-phenyl)propane-1,3-diol, (1S,2R)-1,2-bis(3-methoxy-4-oxidanyl-phenyl)propane-1,3-diol, GLYCEROL, ... | | 著者 | Zahn, M, Kuatsjah, E, Beckham, G.T, McGeehan, J.E. | | 登録日 | 2022-07-04 | | 公開日 | 2023-02-01 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.683 Å) | | 主引用文献 | Biochemical and structural characterization of a sphingomonad diarylpropane lyase for cofactorless deformylation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ABT
 
 | | Crystal structure of NaLdpA in complex with the product analog Resveratrol | | 分子名称: | RESVERATROL, SULFATE ION, SnoaL-like domain-containing protein | | 著者 | Zahn, M, Kuatsjah, E, Beckham, G.T, McGeehan, J.E. | | 登録日 | 2022-07-04 | | 公開日 | 2023-02-01 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | Biochemical and structural characterization of a sphingomonad diarylpropane lyase for cofactorless deformylation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ABU
 
 | | Crystal structure of NaLdpA mutant H97Q in complex with erythro-DGPD | | 分子名称: | (1S,2R)-1,2-bis(3-methoxy-4-oxidanyl-phenyl)propane-1,3-diol, SnoaL-like domain-containing protein | | 著者 | Zahn, M, Kuatsjah, E, Beckham, G.T, McGeehan, J.E. | | 登録日 | 2022-07-04 | | 公開日 | 2023-02-01 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.661 Å) | | 主引用文献 | Biochemical and structural characterization of a sphingomonad diarylpropane lyase for cofactorless deformylation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ABW
 
 | | Crystal structure of SpLdpA in complex with threo-DGPD | | 分子名称: | (1R,2R)-1,2-bis(3-methoxy-4-oxidanyl-phenyl)propane-1,3-diol, (1S,2S)-1,2-bis(3-methoxy-4-oxidanyl-phenyl)propane-1,3-diol, SULFATE ION, ... | | 著者 | Zahn, M, Kuatsjah, E, Beckham, G.T, McGeehan, J.E. | | 登録日 | 2022-07-04 | | 公開日 | 2023-02-01 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Biochemical and structural characterization of a sphingomonad diarylpropane lyase for cofactorless deformylation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6M63
 
 | | Crystal structure of a cAMP sensor G-Flamp1. | | 分子名称: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Chimera of Cyclic nucleotide-gated potassium channel mll3241 and Yellow fluorescent protein | | 著者 | Zhou, Z, Chen, S, Wang, L, Chu, J. | | 登録日 | 2020-03-12 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | A high-performance genetically encoded fluorescent indicator for in vivo cAMP imaging.
Nat Commun, 13, 2022
|
|
5YE9
 
 | | The crystal structure of Lp-PLA2 in complex with a novel inhibitor | | 分子名称: | N-[4-[(3-cyano-4-naphthalen-2-yloxy-phenyl)sulfamoyl]phenyl]ethanamide, Platelet-activating factor acetylhydrolase, SULFATE ION | | 著者 | Liu, Q.F, Xu, Y.C. | | 登録日 | 2017-09-15 | | 公開日 | 2018-07-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.876 Å) | | 主引用文献 | Structure-Guided Discovery of Novel, Potent, and Orally Bioavailable Inhibitors of Lipoprotein-Associated Phospholipase A2.
J. Med. Chem., 60, 2017
|
|
5YE7
 
 | | The crystal structure of Lp-PLA2 in complex with a novel inhibitor | | 分子名称: | N-[4-[(4-naphthalen-2-yloxyphenyl)sulfamoyl]phenyl]ethanamide, Platelet-activating factor acetylhydrolase, SULFATE ION | | 著者 | Liu, Q.F, Xu, Y.C. | | 登録日 | 2017-09-15 | | 公開日 | 2018-07-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.312 Å) | | 主引用文献 | Structure-Guided Discovery of Novel, Potent, and Orally Bioavailable Inhibitors of Lipoprotein-Associated Phospholipase A2.
J. Med. Chem., 60, 2017
|
|
5YE8
 
 | |
5YEA
 
 | | The crystal structure of Lp-PLA2 in complex with a novel inhibitor | | 分子名称: | 4-[[4-[4-chloranyl-3-(trifluoromethyl)phenoxy]-3-cyano-phenyl]sulfamoyl]benzoic acid, Platelet-activating factor acetylhydrolase, SULFATE ION | | 著者 | Liu, Q.F, Xu, Y.C. | | 登録日 | 2017-09-15 | | 公開日 | 2018-07-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.805 Å) | | 主引用文献 | Structure-Guided Discovery of Novel, Potent, and Orally Bioavailable Inhibitors of Lipoprotein-Associated Phospholipase A2.
J. Med. Chem., 60, 2017
|
|
8GDS
 
 | |
1D9X
 
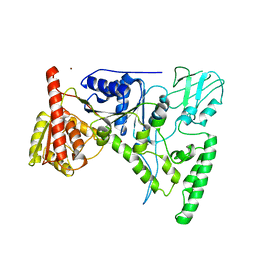 | | CRYSTAL STRUCTURE OF THE DNA REPAIR PROTEIN UVRB | | 分子名称: | EXCINUCLEASE UVRABC COMPONENT UVRB, ZINC ION | | 著者 | Theis, K, Chen, P.J, Skorvaga, M, Van Houten, B, Kisker, C. | | 登録日 | 1999-10-30 | | 公開日 | 2000-05-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of UvrB, a DNA helicase adapted for nucleotide excision repair.
EMBO J., 18, 1999
|
|
1D9Z
 
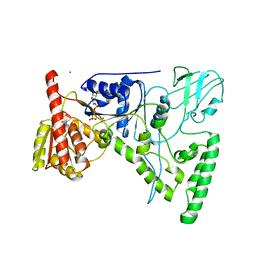 | | CRYSTAL STRUCTURE OF THE DNA REPAIR PROTEIN UVRB IN COMPLEX WITH ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, EXCINUCLEASE UVRABC COMPONENT UVRB, MAGNESIUM ION, ... | | 著者 | Theis, K, Chen, P.J, Skorvaga, M, Van Houten, B, Kisker, C. | | 登録日 | 1999-10-30 | | 公開日 | 2000-05-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Crystal structure of UvrB, a DNA helicase adapted for nucleotide excision repair.
EMBO J., 18, 1999
|
|
8SJQ
 
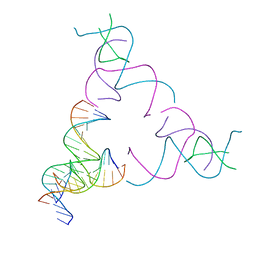 | | [3T16] Self-assembling right-handed tensegrity triangle with 16 interjunction base pairs and R3 symmetry | | 分子名称: | DNA (5'-D(*CP*AP*GP*CP*AP*TP*GP*CP*CP*TP*GP*AP*TP*AP*CP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*CP*TP*GP*TP*GP*GP*CP*AP*TP*GP*C)-3'), DNA (5'-D(P*TP*CP*GP*TP*GP*GP*AP*CP*AP*GP*CP*G)-3'), ... | | 著者 | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | 登録日 | 2023-04-18 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (6.19 Å) | | 主引用文献 | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJR
 
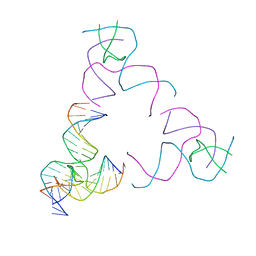 | | [3T17] Self-assembling right-handed tensegrity triangle with 17 interjunction base pairs and R3 symmetry | | 分子名称: | DNA (5'-D(*CP*AP*GP*CP*AP*GP*CP*CP*TP*GP*AP*AP*TP*AP*CP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*CP*TP*GP*TP*GP*GP*CP*TP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*GP*TP*AP*TP*TP*CP*AP*CP*CP*AP*CP*GP*AP*T)-3'), ... | | 著者 | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | 登録日 | 2023-04-18 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (5.25 Å) | | 主引用文献 | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJU
 
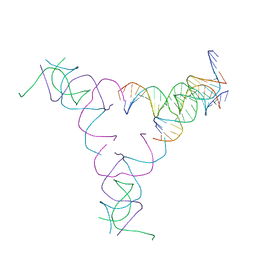 | | [4T17] Self-assembling right-handed four-turn tensegrity triangle with 17 interjunction base pairs and R3 symmetry | | 分子名称: | DNA (25-MER), DNA (5'-D(*GP*AP*AP*AP*AP*AP*CP*AP*CP*TP*GP*CP*CP*TP*GP*AP*AP*TP*AP*CP*CP*GP*CP*A)-3'), DNA (5'-D(P*GP*CP*GP*GP*TP*AP*TP*TP*CP*AP*CP*CP*AP*CP*GP*AP*T)-3'), ... | | 著者 | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | 登録日 | 2023-04-18 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (7.14 Å) | | 主引用文献 | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJM
 
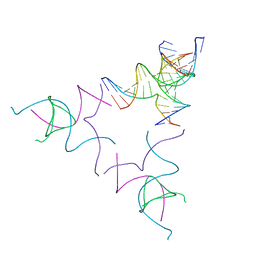 | | [3T12] Self-assembling left-handed tensegrity triangle with 12 interjunction base pairs and R3 symmetry | | 分子名称: | DNA (5'-D(*CP*AP*GP*CP*AP*TP*CP*GP*CP*CP*TP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*CP*GP*CP*AP*TP*GP*TP*GP*GP*CP*GP*AP*TP*GP*C)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*AP*CP*AP*TP*GP*CP*GP*AP*G)-3'), ... | | 著者 | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | 登録日 | 2023-04-18 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (8.08 Å) | | 主引用文献 | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJO
 
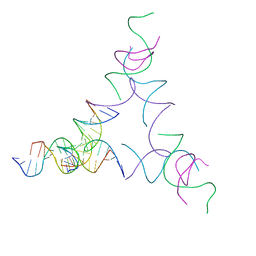 | | [3T14] Self-assembling left-handed tensegrity triangle with 14 interjunction base pairs and R3 symmetry | | 分子名称: | DNA (5'-D(P*CP*AP*CP*GP*TP*GP*GP*AP*CP*AP*GP*GP*AP*G)-3'), DNA (5'-D(P*CP*AP*GP*CP*TP*CP*AP*GP*CP*CP*TP*GP*AP*CP*TP*CP*A)-3'), DNA (5'-D(P*GP*TP*GP*AP*GP*TP*CP*TP*CP*CP*AP*CP*GP*T)-3'), ... | | 著者 | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | 登録日 | 2023-04-18 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (7.08 Å) | | 主引用文献 | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJN
 
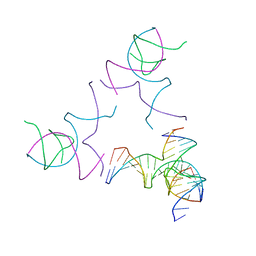 | | [3T13] Self-assembling left-handed tensegrity triangle with 13 interjunction base pairs and R3 symmetry | | 分子名称: | DNA (5'-D(*CP*AP*GP*CP*AP*TP*CP*GP*CP*CP*TP*GP*AP*CP*TP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*CP*GP*CP*TP*GP*TP*GP*GP*CP*GP*AP*TP*GP*C)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*AP*CP*AP*GP*CP*GP*AP*G)-3'), ... | | 著者 | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | 登録日 | 2023-04-18 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (7.3 Å) | | 主引用文献 | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJP
 
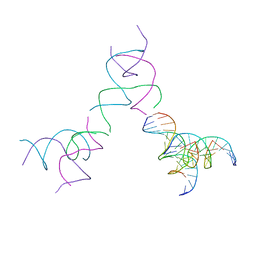 | | [3T15] Self-assembling DNA motif with 15 base pairs between junctions and P32 symmetry | | 分子名称: | DNA (5'-D(*CP*AP*GP*CP*TP*GP*AP*CP*CP*TP*GP*AP*CP*TP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*CP*CP*TP*GP*TP*GP*GP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*CP*GP*AP*TP*GP*GP*AP*CP*AP*GP*GP*GP*G)-3'), ... | | 著者 | Vecchioni, S, Janowski, J, Sha, R, Ohayon, Y.P. | | 登録日 | 2023-04-18 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (5.22 Å) | | 主引用文献 | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJW
 
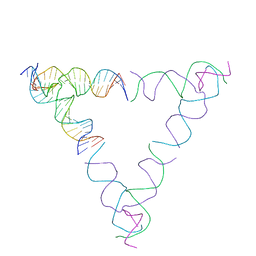 | | [4T28] Self-assembling right-handed tensegrity triangle with 28 interjunction base pairs and R3 symmetry | | 分子名称: | DNA (28-MER), DNA (5'-D(*GP*AP*AP*CP*TP*GP*CP*CP*TP*GP*AP*AP*TP*TP*AP*CP*TP*GP*AP*CP*CP*G)-3'), DNA (5'-D(*TP*CP*AP*TP*CP*AP*GP*TP*GP*GP*CP*AP*GP*T)-3'), ... | | 著者 | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | 登録日 | 2023-04-18 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (7.64 Å) | | 主引用文献 | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
